Notice (8): Undefined variable: solution_of_interest [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'product' => array(
'Product' => array(
'id' => '1979',
'antibody_id' => '631',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (sample size)',
'description' => '<p>Monoclonal antibody raised in mouse against 5-mC (<strong>5-methylcytosine</strong>) conjugated to ovalbumine (<strong>33D3 clone</strong>).</p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15200081-10',
'old_catalog_number' => 'MAb-081-010',
'sf_code' => 'C15200081-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10-ug-5-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:58:36',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '631',
'name' => '5-mC monoclonal antibody 33D3 (sample size)',
'description' => '',
'clonality' => '',
'isotype' => 'IgG1',
'lot' => 'RD-006',
'concentration' => '1.1 µg/µl',
'reactivity' => 'Human, mouse, rat, cow, other (wide range): positive',
'type' => 'Monoclonal. <br /><strong>MeDIP grade.</strong><br /><strong>MeDIP-seq grade</strong>.',
'purity' => 'Protein A purified monoclonal antibody.',
'classification' => '',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>MeDIP/MeDIP-seq <sup>*</sup></td>
<td>0.1 - 2 µg/IP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting **</td>
<td>1:300</td>
<td>Fig 3</td>
</tr>
<tr>
<td>IF</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
</tbody>
</table>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.1-5 µg per IP.</small></p>
<p><small><sup>**</sup> Dot blot was only performed to demonstrate the specificity. This antibody is not recommended for dot blot on biological samples.</small></p>
<p></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2023-05-11 15:21:00',
'created' => '2020-12-17 10:31:21',
'select_label' => '631 - 5-mC monoclonal antibody 33D3 (sample size) (RD-006 - 1.1 µg/µl - Human, mouse, rat, cow, other (wide range): positive - Protein A purified monoclonal antibody. - Mouse)'
),
'Slave' => array(),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
),
(int) 25 => array(
[maximum depth reached]
),
(int) 26 => array(
[maximum depth reached]
),
(int) 27 => array(
[maximum depth reached]
),
(int) 28 => array(
[maximum depth reached]
),
(int) 29 => array(
[maximum depth reached]
),
(int) 30 => array(
[maximum depth reached]
),
(int) 31 => array(
[maximum depth reached]
),
(int) 32 => array(
[maximum depth reached]
),
(int) 33 => array(
[maximum depth reached]
),
(int) 34 => array(
[maximum depth reached]
),
(int) 35 => array(
[maximum depth reached]
),
(int) 36 => array(
[maximum depth reached]
),
(int) 37 => array(
[maximum depth reached]
),
(int) 38 => array(
[maximum depth reached]
),
(int) 39 => array(
[maximum depth reached]
),
(int) 40 => array(
[maximum depth reached]
),
(int) 41 => array(
[maximum depth reached]
),
(int) 42 => array(
[maximum depth reached]
),
(int) 43 => array(
[maximum depth reached]
),
(int) 44 => array(
[maximum depth reached]
),
(int) 45 => array(
[maximum depth reached]
),
(int) 46 => array(
[maximum depth reached]
),
(int) 47 => array(
[maximum depth reached]
),
(int) 48 => array(
[maximum depth reached]
),
(int) 49 => array(
[maximum depth reached]
),
(int) 50 => array(
[maximum depth reached]
),
(int) 51 => array(
[maximum depth reached]
),
(int) 52 => array(
[maximum depth reached]
),
(int) 53 => array(
[maximum depth reached]
),
(int) 54 => array(
[maximum depth reached]
),
(int) 55 => array(
[maximum depth reached]
),
(int) 56 => array(
[maximum depth reached]
),
(int) 57 => array(
[maximum depth reached]
),
(int) 58 => array(
[maximum depth reached]
),
(int) 59 => array(
[maximum depth reached]
),
(int) 60 => array(
[maximum depth reached]
),
(int) 61 => array(
[maximum depth reached]
),
(int) 62 => array(
[maximum depth reached]
),
(int) 63 => array(
[maximum depth reached]
),
(int) 64 => array(
[maximum depth reached]
),
(int) 65 => array(
[maximum depth reached]
),
(int) 66 => array(
[maximum depth reached]
),
(int) 67 => array(
[maximum depth reached]
),
(int) 68 => array(
[maximum depth reached]
),
(int) 69 => array(
[maximum depth reached]
),
(int) 70 => array(
[maximum depth reached]
),
(int) 71 => array(
[maximum depth reached]
),
(int) 72 => array(
[maximum depth reached]
),
(int) 73 => array(
[maximum depth reached]
),
(int) 74 => array(
[maximum depth reached]
),
(int) 75 => array(
[maximum depth reached]
),
(int) 76 => array(
[maximum depth reached]
),
(int) 77 => array(
[maximum depth reached]
),
(int) 78 => array(
[maximum depth reached]
),
(int) 79 => array(
[maximum depth reached]
),
(int) 80 => array(
[maximum depth reached]
),
(int) 81 => array(
[maximum depth reached]
),
(int) 82 => array(
[maximum depth reached]
),
(int) 83 => array(
[maximum depth reached]
),
(int) 84 => array(
[maximum depth reached]
),
(int) 85 => array(
[maximum depth reached]
),
(int) 86 => array(
[maximum depth reached]
),
(int) 87 => array(
[maximum depth reached]
),
(int) 88 => array(
[maximum depth reached]
),
(int) 89 => array(
[maximum depth reached]
),
(int) 90 => array(
[maximum depth reached]
),
(int) 91 => array(
[maximum depth reached]
),
(int) 92 => array(
[maximum depth reached]
),
(int) 93 => array(
[maximum depth reached]
),
(int) 94 => array(
[maximum depth reached]
),
(int) 95 => array(
[maximum depth reached]
),
(int) 96 => array(
[maximum depth reached]
),
(int) 97 => array(
[maximum depth reached]
),
(int) 98 => array(
[maximum depth reached]
),
(int) 99 => array(
[maximum depth reached]
),
(int) 100 => array(
[maximum depth reached]
),
(int) 101 => array(
[maximum depth reached]
),
(int) 102 => array(
[maximum depth reached]
),
(int) 103 => array(
[maximum depth reached]
),
(int) 104 => array(
[maximum depth reached]
),
(int) 105 => array(
[maximum depth reached]
),
(int) 106 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul'
)
$language = 'en'
$meta_keywords = ''
$meta_description = '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.'
$meta_title = '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode'
$product = array(
'Product' => array(
'id' => '1979',
'antibody_id' => '631',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (sample size)',
'description' => '<p>Monoclonal antibody raised in mouse against 5-mC (<strong>5-methylcytosine</strong>) conjugated to ovalbumine (<strong>33D3 clone</strong>).</p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
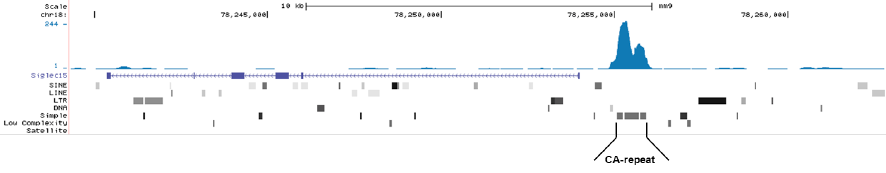
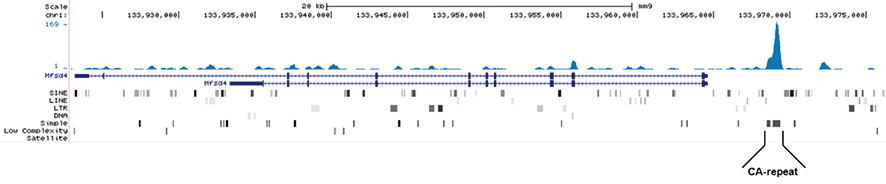
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
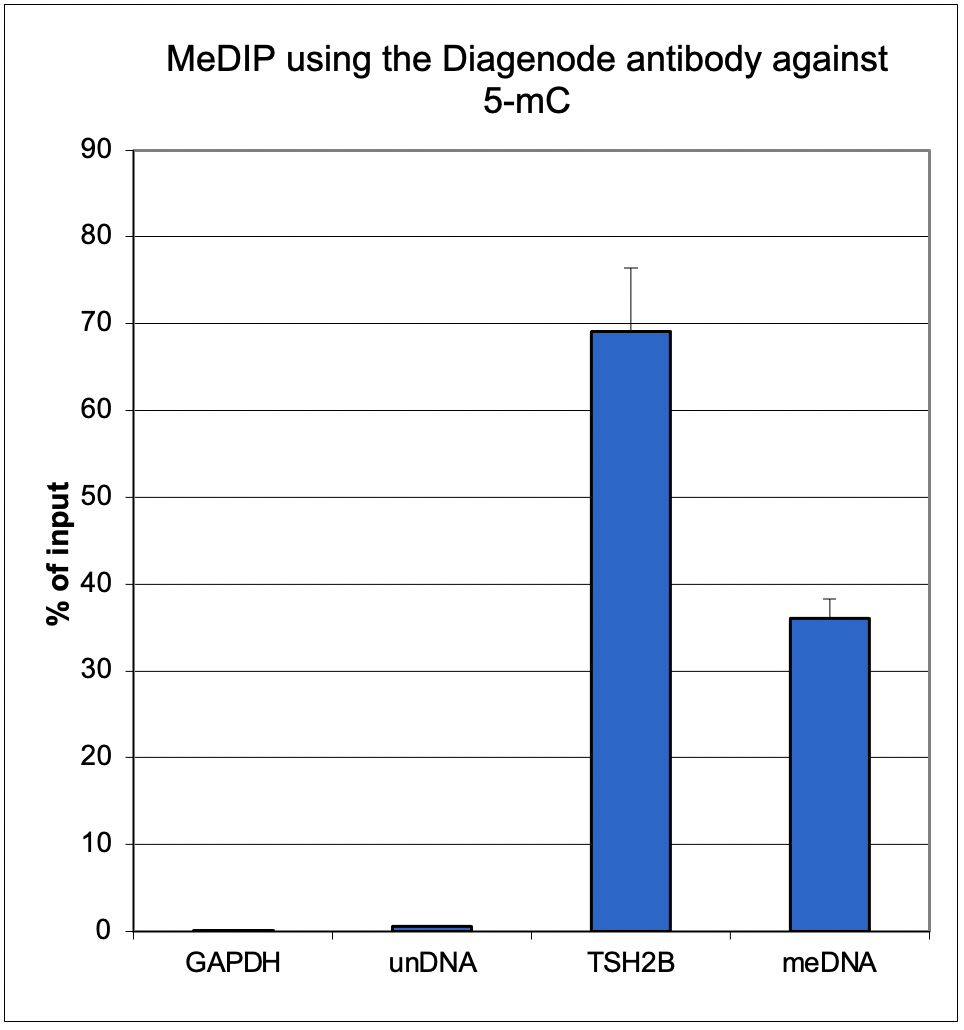
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
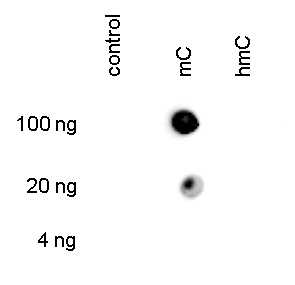
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
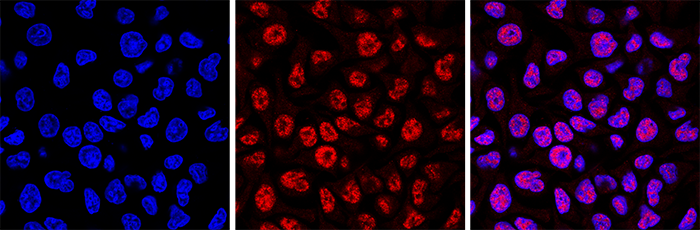
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15200081-10',
'old_catalog_number' => 'MAb-081-010',
'sf_code' => 'C15200081-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10-ug-5-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:58:36',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '631',
'name' => '5-mC monoclonal antibody 33D3 (sample size)',
'description' => '',
'clonality' => '',
'isotype' => 'IgG1',
'lot' => 'RD-006',
'concentration' => '1.1 µg/µl',
'reactivity' => 'Human, mouse, rat, cow, other (wide range): positive',
'type' => 'Monoclonal. <br /><strong>MeDIP grade.</strong><br /><strong>MeDIP-seq grade</strong>.',
'purity' => 'Protein A purified monoclonal antibody.',
'classification' => '',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>MeDIP/MeDIP-seq <sup>*</sup></td>
<td>0.1 - 2 µg/IP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting **</td>
<td>1:300</td>
<td>Fig 3</td>
</tr>
<tr>
<td>IF</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
</tbody>
</table>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.1-5 µg per IP.</small></p>
<p><small><sup>**</sup> Dot blot was only performed to demonstrate the specificity. This antibody is not recommended for dot blot on biological samples.</small></p>
<p></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2023-05-11 15:21:00',
'created' => '2020-12-17 10:31:21',
'select_label' => '631 - 5-mC monoclonal antibody 33D3 (sample size) (RD-006 - 1.1 µg/µl - Human, mouse, rat, cow, other (wide range): positive - Protein A purified monoclonal antibody. - Mouse)'
),
'Slave' => array(),
'Group' => array(
'Group' => array(
'id' => '4',
'name' => 'C15200081',
'product_id' => '1980',
'modified' => '2016-02-17 17:32:42',
'created' => '2016-02-17 17:32:42'
),
'Master' => array(
'id' => '1980',
'antibody_id' => '630',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>Monoclonal antibody raised in mouse against </span><b>5-mC</b><span><span> </span>(</span><b>5-methylcytosine</b><span>) conjugated to ovalbumine (</span><b>33D3 clone</b><span>).</span></p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15200081-100',
'old_catalog_number' => 'MAb-081-100',
'sf_code' => 'C15200081-D001-000526',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '505',
'price_USD' => '575',
'price_GBP' => '450',
'price_JPY' => '79110',
'price_CNY' => '0',
'price_AUD' => '1438',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'October 27, 2020',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081) | Diagenode',
'meta_keywords' => '5-methylcytosine (5-mC),monoclonal antibody,Methylated DNA Immunoprecipitation',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2023-05-17 10:08:33',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1885',
'antibody_id' => null,
'name' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'description' => '<p><span>The Auro hMeDIP kit is designed for enrichment of hydroxymethylated DNA from fragmented genomic DNA samples for use in genome-wide methylation analysis. It features</span><span> a highly specific monoclonal antibody against </span><span>5-hydroxymethylcytosine (5-hmC) for the immunoprecipitation of hydroxymethylated DNA</span><span>. It includes control DNA and primers to assess the effiency of the assay. </span><span>Performing hydroxymethylation profiling with the hMeDIP kit is fast, reliable and highly specific.</span></p>',
'label1' => ' Characteristics',
'info1' => '<ul style="list-style-type: disc;">
<li><span>Robust enrichment & immunoprecipitation of hydroxymethylated DNA</span></li>
<li>Highly specific monoclonal antibody against 5-hmC<span> for reliable, reproducible results</span></li>
<li>Including control DNA and primers to <span>monitor the efficiency of the assay</span>
<ul style="list-style-type: circle;">
<li>5-hmC, 5-mC and unmethylated DNA sequences and primer pairs</li>
<li>Mouse primer pairs against Sfi1 targeting hydroxymethylated gene in mouse</li>
</ul>
</li>
</ul>
<ul style="list-style-type: disc;">
<li>Improved single-tube, magnetic bead-based protocol</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C02010034',
'old_catalog_number' => '',
'sf_code' => 'C02010034-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '630',
'price_USD' => '690',
'price_GBP' => '580',
'price_JPY' => '98690',
'price_CNY' => '',
'price_AUD' => '1725',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns',
'meta_title' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'meta_keywords' => '',
'meta_description' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'modified' => '2021-01-18 10:37:19',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1880',
'antibody_id' => null,
'name' => 'MagMeDIP Kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/magmedip-kit-manual-C02010020-21.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p> </p>
<div class="small-12 medium-4 large-4 columns"><center></center><center></center><center></center><center><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank"><img src="https://www.diagenode.com/img/banners/banner-nature-publication-580.png" alt="Click here to read more about MeDIP " caption="false" width="80%" /></a></center></div>
<div class="small-12 medium-8 large-8 columns">
<h3 style="text-align: justify;">Sensitive tumour detection and classification using plasma cell-free DNA methylomes<br /><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank">Read the publication</a></h3>
<h3 class="c-article-title u-h1" data-test="article-title" itemprop="name headline" style="text-align: justify;">Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA<br /><a href="https://www.nature.com/articles/s41596-019-0202-2" target="_blank" title="cfMeDIP-seq Nature Method">Read the method</a></h3>
</div>
<p></p>
<p></p>
<p></p>
<div class="row">
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
<div class="small-12 medium-8 large-8 columns"><br />
<p>Perform <strong>MeDIP</strong> (<strong>Me</strong>thylated <strong>D</strong>NA <strong>I</strong>mmuno<strong>p</strong>recipitation) followed by qPCR or NGS to estimate DNA methylation status of your sample using a highly sensitive 5-methylcytosine antibody. Our MagMeDIP kit contains high quality reagents to get the highest enrichment of methylated DNA with an optimized user-friendly protocol.</p>
</div>
</div>
<h3><span>Features</span></h3>
<ul>
<li>Starting DNA amount: <strong>10 ng – 1 µg</strong></li>
<li>Content: <strong>all reagents included</strong> for DNA extraction, immunoprecipitation (including the 5-mC antibody, spike-in controls and their corresponding qPCR primer pairs) as well as DNA isolation after IP.</li>
<li>Application: <strong>qPCR</strong> and <strong>NGS</strong></li>
<li>Robust method, <strong>superior enrichment</strong>, and easy-to-use protocol</li>
<li><strong>High reproducibility</strong> between replicates and repetitive experiments</li>
<li>Compatible with <strong>all species </strong></li>
</ul>',
'label1' => 'MagMeDIP workflow',
'info1' => '<p>DNA methylation occurs primarily as 5-methylcytosine (5-mC), and the Diagenode MagMeDIP Kit takes advantage of a specific antibody targeting this 5-mC to immunoprecipitate methylated DNA, which can be thereafter directly analyzed by qPCR or Next-Generation Sequencing (NGS).</p>
<h3><span>How it works</span></h3>
<p>In brief, after the cell collection and lysis, the genomic DNA is extracted, sheared, and then denatured. In the next step the antibody directed against 5 methylcytosine and antibody binding beads are used for immunoselection and immunoprecipitation of methylated DNA fragments. Then, the IP’d methylated DNA is isolated and can be used for any subsequent analysis as qPCR, amplification, hybridization on microarrays or next generation sequencing.</p>
<center><img src="https://www.diagenode.com/img/product/kits/MagMeDIP-workflow.png" width="70%" alt="5-methylcytosine" caption="false" /></center>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>',
'label2' => 'MeDIP-qPCR',
'info2' => '<p>The kit MagMeDIP contains all reagents necessary for a complete MeDIP-qPCR workflow. Two MagMeDIP protocols have been validated: for manual processing as well as for automated processing, using the Diagenode’s IP-Star Compact Automated System (please refer to the kit manual).</p>
<ul>
<li><strong>Complete kit</strong> including DNA extraction module, IP antibody and reagents, DNA isolation buffer</li>
<li><strong>Quality control of the IP:</strong> due to methylated and unmethylated DNA spike-in controls and their associated qPCR primers</li>
<li><strong>Easy to use</strong> with user-friendly magnetic beads and rack</li>
<li><strong>Highly validated protocol</strong></li>
<li>Automated protocol supplied</li>
</ul>
<center><img src="https://www.diagenode.com/img/product/kits/fig1-magmedipkit.png" width="85%" alt="Methylated DNA Immunoprecipitation" caption="false" /></center>
<p style="font-size: 0.9em;"><em><strong>Figure 1.</strong> Immunoprecipitation results obtained with Diagenode MagMeDIP Kit</em></p>
<p style="font-size: 0.9em;">MeDIP assays were performed manually using 1 µg or 50 ng gDNA from blood cells with the MagMeDIP kit (Diagenode). The IP was performed with the Methylated and Unmethylated spike-in controls included in the kit, together with the human DNA samples. The DNA was isolated/purified using DIB. Afterwards, qPCR was performed using the primer pairs included in this kit.</p>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>',
'label3' => 'MeDIP-seq',
'info3' => '<p>For DNA methylation analysis on the whole genome, MagMeDIP kit can be coupled with Next-Generation Sequencing. To perform MeDIP-sequencing we recommend the following strategy:</p>
<ul style="list-style-type: circle;">
<li>Choose a library preparation solution which is compatible with the starting amount of DNA you are planning to use (from 10 ng to 1 μg). It can be a home-made solution or a commercial one.</li>
<li>Choose the indexing system that fits your needs considering the following features:</li>
<ul>
<ul>
<ul>
<li>Single-indexing, combinatorial dual-indexing or unique dual-indexing</li>
<li>Number of barcodes</li>
<li>Full-length adaptors containing the barcodes or barcoding at the final amplification step</li>
<li>Presence / absence of Unique Molecular Identifiers (for PCR duplicates removal)</li>
</ul>
</ul>
</ul>
<li>Standard library preparation protocols are compatible with double-stranded DNA only, therefore the first steps of the library preparation (end repair, A-tailing, adaptor ligation and clean-up) will have to be performed on sheared DNA, before the IP.</li>
</ul>
<p style="padding-left: 30px;"><strong>CAUTION:</strong> As the immunoprecipitation step occurs at the middle of the library preparation workflow, single-tube solutions for library preparation are usually not compatible with MeDIP-sequencing.</p>
<ul style="list-style-type: circle;">
<li>For DNA isolation after the IP, we recommend using the <a href="https://www.diagenode.com/en/p/ipure-kit-v2-x24" title="IPure kit v2">IPure kit v2</a> (available separately, Cat. No. C03010014) instead of DNA isolation Buffer.</li>
</ul>
<ul style="list-style-type: circle;">
<li>Perform library amplification after the DNA isolation following the standard protocol of the chosen library preparation solution.</li>
</ul>
<h3><span>MeDIP-seq workflow</span></h3>
<center><img src="https://www.diagenode.com/img/product/kits/MeDIP-seq-workflow.png" width="110%" alt="MagMeDIP qPCR Kit x10 workflow" caption="false" /></center>
<h3><span>Example of results</span></h3>
<center><img src="https://www.diagenode.com/img/product/kits/medip-specificity.png" alt="MagMeDIP qPCR Kit Result" caption="false" width="951" height="488" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 1. qPCR analysis of external spike-in DNA controls (methylated and unmethylated) after IP.</strong> Samples were prepared using 1μg – 100ng -10ng sheared human gDNA with the MagMeDIP kit (Diagenode) and a commercially available library prep kit. DNA isolation after IP has been performed with IPure kit V2 (Diagenode).</p>
<p></p>
<p></p>
<center><img src="https://www.diagenode.com/img/product/kits/medip-saturation-analysis.png" alt=" MagMeDIP kit " caption="false" width="951" height="461" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 2. Saturation analysis.</strong> Clean reads were aligned to the human genome (hg19) using Burrows-Wheeler aligner (BWA) algorithm after which duplicated and unmapped reads were removed resulting in a mapping efficiency >98% for all samples. Quality and validity check of the mapped MeDIP-seq data was performed using MEDIPS R package. Saturation plots show that all sets of reads have sufficient complexity and depth to saturate the coverage profile of the reference genome and that this is reproducible between replicates and repetitive experiments (data shown for 50 ng gDNA input: left panel = replicate a, right panel = replicate b).</p>
<p></p>
<p></p>
<center><img src="https://www.diagenode.com/img/product/kits/medip-libraries-prep.png" alt="MagMeDIP x10 " caption="false" width="951" height="708" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 3. Sequencing profiles of MeDIP-seq libraries prepared from different starting amounts of sheared gDNA on the positive and negative methylated control regions.</strong> MeDIP-seq libraries were prepared from decreasing starting amounts of gDNA (1 μg (green), 50 ng (red), and 10ng (blue)) originating from human blood with the MagMeDIP kit (Diagenode) and a commercially available library prep kit. DNA isolation after IP has been performed with IPure kit V2 (Diagenode). IP and corresponding INPUT samples were sequenced on Illumina NovaSeq SP with 2x50 PE reads. The reads were mapped to the human genome (hg19) with bwa and the alignments were loaded into IGV (the tracks use an identical scale). The top IGV figure shows the TSH2B (also known as H2BC1) gene (marked by blue boxes in the bottom track) and its surroundings. The TSH2B gene is coding for a histone variant that does not occur in blood cells, and it is known to be silenced by methylation. Accordingly, we see a high coverage in the vicinity of this gene. The bottom IGV figure shows the GADPH locus (marked by blue boxes in the bottom track) and its surroundings. The GADPH gene is a highly active transcription region and should not be methylated, resulting in no reads accumulation following MeDIP-seq experiment.</p>
<p></p>
<ul>
<ul>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
</ul>
</ul>',
'format' => '48 rxns (IP)',
'catalog_number' => 'C02010021',
'old_catalog_number' => 'mc-magme-048',
'sf_code' => 'C02010021-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '745',
'price_USD' => '750',
'price_GBP' => '680',
'price_JPY' => '116705',
'price_CNY' => '',
'price_AUD' => '1875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'magmedip-kit-x48-48-rxns',
'meta_title' => 'MagMeDIP Kit for efficient immunoprecipitation of methylated DNA | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Perform Methylated DNA Immunoprecipitation (MeDIP) to estimate DNA methylation status of your sample using highly specific 5-mC antibody. This kit allows the preparation of cfMeDIP-seq libraries.',
'modified' => '2023-07-06 14:15:10',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1887',
'antibody_id' => null,
'name' => 'MethylCap kit',
'description' => '<p>The MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. The procedure has been adapted to both manual process or <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a>. Libraries of captured methylated DNA can be prepared for next-generation sequencing (NGS) by combining MBD technology with the <a href="https://www.diagenode.com/en/p/microplex-lib-prep-kit-v3-48-rxns">MicroPlex Library Preparation Kit v3</a>.</p>',
'label1' => 'Characteristics',
'info1' => '<ul style="list-style-type: circle;">
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>On-day protocol</strong></li>
<li><strong>NGS compatibility</strong></li>
</ul>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong></strong></p>
<p><strong></strong><strong>F</strong><strong>igure 1.</strong> Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).</p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020010',
'old_catalog_number' => 'AF-100-0048',
'sf_code' => 'C02020010-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'methylcap-kit-x48-48-rxns',
'meta_title' => 'MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'MethylCap kit x48',
'modified' => '2021-02-19 17:55:38',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '1892',
'antibody_id' => null,
'name' => 'Premium Bisulfite kit',
'description' => '<p style="text-align: center;"><a href="https://www.diagenode.com/files/products/kits/Premium_Bisulfite_kit_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p style="text-align: center;"><strong>Make your Bisulfite conversion now in only 60 minutes !</strong></p>
<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C02030030',
'old_catalog_number' => '',
'sf_code' => 'C02030030-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'premium-bisulfite-kit-50-rxns',
'meta_title' => 'Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Premium Bisulfite kit',
'modified' => '2023-04-20 16:13:50',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array([maximum depth reached])
)
),
'Application' => array(
(int) 0 => array(
'id' => '28',
'position' => '10',
'parent_id' => '40',
'name' => 'DB',
'description' => '<p>Dot blotting</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'dot-blotting',
'meta_keywords' => 'Dot blotting,Monoclonal & Polyclonal antibody,',
'meta_description' => 'Diagenode offers Monoclonal & Polyclonal antibodies for Dot blotting applications',
'meta_title' => 'Dot blotting Antibodies - Monoclonal & Polyclonal antibody | Diagenode',
'modified' => '2016-01-13 14:40:49',
'created' => '2015-07-08 13:45:05',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '23',
'position' => '40',
'parent_id' => '4',
'name' => 'DNA modifications',
'description' => '<p><span style="font-weight: 400;">T</span><span style="font-weight: 400;">he pattern of <strong>DNA modifications</strong> is critical for genome stability and the control of gene expression in the cell. Methylation of 5-cytosine (5-mC), one of the best-studied epigenetic marks, is carried out by the <strong>DNA methyltransferases</strong> DNMT3A and B and DNMT1. DNMT3A and DNMT3B are responsible for </span><i><span style="font-weight: 400;">de novo</span></i><span style="font-weight: 400;"> DNA methylation, whereas DNMT1 maintains existing methylation. 5-mC undergoes active demethylation which is performed by the <strong>Ten-Eleven Translocation</strong> (TET) familly of DNA hydroxylases. The latter consists of 3 members TET1, 2 and 3. All 3 members catalyze the conversion of <strong>5-methylcytosine</strong> (5-mC) into <strong>5-hydroxymethylcytosine</strong> (5-hmC), and further into <strong>5-formylcytosine</strong> (5-fC) and <strong>5-carboxycytosine</strong> (5-caC). 5-fC and 5-caC can be converted to unmodified cytosine by <strong>Thymine DNA Glycosylase</strong> (TDG). It is not yet clear if 5-hmC, 5-fC and 5-caC have specific functions or are simply intermediates in the demethylation of 5-mC.</span></p>
<p><span style="font-weight: 400;">DNA methylation is generally considered as a repressive mark and is usually associated with gene silencing. It is essential that the balance between DNA methylation and demethylation is precisely maintained. Dysregulation of DNA methylation may lead to many different human diseases and is often observed in cancer cells.</span></p>
<p><span style="font-weight: 400;">Diagenode offers highly validated antibodies against different proteins involved in DNA modifications as well as against the modified bases allowing the study of all steps and intermediates in the DNA methylation/demethylation pathway:</span></p>
<p><img src="https://www.diagenode.com/img/categories/antibodies/dna-methylation.jpg" height="599" width="816" /></p>
<p><strong>Diagenode exclusively sources the original 5-methylcytosine monoclonal antibody (clone 33D3).</strong></p>
<p>Check out the list below to see all proposed antibodies for DNA modifications.</p>
<p>Diagenode’s highly validated antibodies:</p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'dna-methylation-antibody',
'cookies_tag_id' => null,
'meta_keywords' => 'Monoclonal Antibodies,Polyclonal antibodies,DNA methylation,Diagenode',
'meta_description' => 'Diagenode offers Monoclonal and Polyclonal antibodies for DNA Methylation. The pattern of DNA modifications is critical for genome stability and the control of gene expression in the cell. ',
'meta_title' => 'DNA modifications - Monoclonal and Polyclonal Antibodies for DNA Methylation | Diagenode',
'modified' => '2020-08-03 15:25:37',
'created' => '2014-07-31 03:51:53',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '102',
'position' => '1',
'parent_id' => '4',
'name' => 'Sample size antibodies',
'description' => '<h1><strong>Validated epigenetics antibodies</strong> – care for a sample?<br /> </h1>
<p>Diagenode has partnered with leading epigenetics experts and numerous epigenetics consortiums to bring to you a validated and comprehensive collection of epigenetic antibodies. As an expert in epigenetics, we are committed to offering highly-specific antibodies validated for ChIP/ChIP-seq and many other applications. All batch-specific validation data is available on our website.<br /><a href="../categories/antibodies">Read about our expertise in antibody production</a>.</p>
<ul>
<li><strong>Focused</strong> - Diagenode's selection of antibodies is exclusively dedicated for epigenetic research. <a title="See the full collection." href="../categories/all-antibodies">See the full collection.</a></li>
<li><strong>Strict quality standards</strong> with rigorous QC and validation</li>
<li><strong>Classified</strong> based on level of validation for flexibility of application</li>
</ul>
<p>Existing sample sizes are listed below. We will soon expand our collection. Are you looking for a sample size of another antibody? Just <a href="mailto:agnieszka.zelisko@diagenode.com?Subject=Sample%20Size%20Request" target="_top">Contact us</a>.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => true,
'is_antibody' => true,
'slug' => 'sample-size-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => '5-hmC monoclonal antibody,CRISPR/Cas9 polyclonal antibody ,H3K36me3 polyclonal antibody,diagenode',
'meta_description' => 'Diagenode offers sample volume on selected antibodies for researchers to test, validate and provide confidence and flexibility in choosing from our wide range of antibodies ',
'meta_title' => 'Sample-size Antibodies | Diagenode',
'modified' => '2019-07-03 10:57:05',
'created' => '2015-10-27 12:13:34',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '15',
'name' => 'Monoclonal Antibody hMeDIP Kit for DNA Hydroxymethylation Studies',
'description' => '<p><span>There is substantial interest and speculation in the role of the “sixth DNA base,” 5-hydroxymethylcytosine (5-hmC), although its precise function has not yet been elucidated. Since its discovery in neuronal Purkinje, granule and ES cells, studies of this new modified DNA base have been limited by the lack of high-quality, validated tools and technologies that discriminate hydroxy-methylation from methylation in regulating genome expression. Obtaining a specific assay for 5-hmC is particularly important since standard bisulfite sequencing cannot distinguish between these two types of methylation. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Monoclonal_Antibody_hMeDIP_Kit_for_DNA_Hydroxymethylation_Studies_Poster.pdf',
'slug' => 'monoclonal-antibody-hmedip-kit-for-dna-hydroxymethylation-studies-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-09-29 13:42:52',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1118',
'name' => 'Datasheet 5-mC33D3 C15200081-10',
'description' => '<p><span>The 5-methylcytosine antibody (clone 33D3) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span></p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_5-mC33D3_C15200081-10.pdf',
'slug' => 'datasheet-5-mc33d3-c15200081-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-12-17 10:55:09',
'created' => '2020-12-17 10:55:09',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '250',
'name' => 'product/antibodies/antibody.png',
'alt' => 'Mouse IgG',
'modified' => '2020-11-27 07:00:09',
'created' => '2015-07-17 10:12:18',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4843',
'name' => 'Differentiation block in acute myeloid leukemia regulated by intronicsequences of FTO',
'authors' => 'Camera F. et al.',
'description' => '<p>Iroquois transcription factor gene IRX3 is highly expressed in 20–30\% of acute myeloid leukemia (AML) and contributes to the pathognomonic differentiation block. Intron 8 FTO sequences ∼220kB downstream of IRX3 exhibit histone acetylation, DNA methylation, and contacts with the IRX3 promoter, which correlate with IRX3 expression. Deletion of these intronic elements confirms a role in positively regulating IRX3. RNAseq revealed long non-coding (lnc) transcripts arising from this locus. FTO-lncAML knockdown (KD) induced differentiation of AML cells, loss of clonogenic activity, and reduced FTO intron 8:IRX3 promoter contacts. While both FTO-lncAML KD and IRX3 KD induced differentiation, FTO-lncAML but not IRX3 KD led to HOXA downregulation suggesting transcript activity in trans. FTO-lncAMLhigh AML samples expressed higher levels of HOXA and lower levels of differentiation genes. Thus, a regulatory module in FTO intron 8 consisting of clustered enhancer elements and a long non-coding RNA is active in human AML, impeding myeloid differentiation.</p>',
'date' => '2023-08-01',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2589004223013962',
'doi' => '10.1016/j.isci.2023.107319',
'modified' => '2023-08-01 14:14:01',
'created' => '2023-08-01 15:59:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4796',
'name' => 'Nicotinamide N-methyltransferase sustains a core epigenetic programthat promotes metastatic colonization in breast cancer.',
'authors' => 'Couto J.P. et al.',
'description' => '<p><span>Metastatic colonization of distant organs accounts for over 90% of deaths related to solid cancers, yet the molecular determinants of metastasis remain poorly understood. Here, we unveil a mechanism of colonization in the aggressive basal-like subtype of breast cancer that is driven by the NAD</span><sup>+</sup><span><span> </span>metabolic enzyme nicotinamide N-methyltransferase (NNMT). We demonstrate that NNMT imprints a basal genetic program into cancer cells, enhancing their plasticity. In line, NNMT expression is associated with poor clinical outcomes in patients with breast cancer. Accordingly, ablation of NNMT dramatically suppresses metastasis formation in pre-clinical mouse models. Mechanistically, NNMT depletion results in a methyl overflow that increases histone H3K9 trimethylation (H3K9me3) and DNA methylation at the promoters of PR/SET Domain-5 (PRDM5) and extracellular matrix-related genes. PRDM5 emerged in this study as a pro-metastatic gene acting via induction of cancer-cell intrinsic transcription of collagens. Depletion of PRDM5 in tumor cells decreases COL1A1 deposition and impairs metastatic colonization of the lungs. These findings reveal a critical activity of the NNMT-PRDM5-COL1A1 axis for cancer cell plasticity and metastasis in basal-like breast cancer.</span></p>',
'date' => '2023-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37259596',
'doi' => '10.15252/embj.2022112559',
'modified' => '2023-06-15 08:35:19',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4787',
'name' => 'The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.',
'authors' => 'Garcia-Gomez E. et al.',
'description' => '<p>Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the disease (control group). In addition, eight of the PCOS patients underwent intervention with metformin (1500 mg/day) and a carbohydrate-controlled diet (type and quantity) for three months. Clinical and metabolic parameters were determined, and RT-qPCR and MeDIP-qPCR were used to evaluate gene expression and DNA methylation levels, respectively. Decreased expression levels of , , and genes and increased DNA methylation levels of the promoter were found in the endometrium of PCOS patients compared to controls. After metformin and nutritional intervention, some metabolic and clinical variables improved in PCOS patients. This intervention was associated with increased expression of , , and genes and reduced DNA methylation levels of the promoter in the endometrium of PCOS women. Our preliminary findings suggest that metformin and a carbohydrate-controlled diet improve endometrial function in PCOS patients, partly by modulating DNA methylation of the gene promoter and the expression of genes implicated in endometrial receptivity and insulin signaling.</p>',
'date' => '2023-04-01',
'pmid' => 'https://doi.org/10.3390%2Fijms24076857',
'doi' => '10.3390/ijms24076857',
'modified' => '2023-06-12 08:58:33',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4777',
'name' => 'Epigenetic modifier alpha-ketoglutarate modulates aberrant gene bodymethylation and hydroxymethylation marks in diabetic heart.',
'authors' => 'Dhat R. et al.',
'description' => '<p>BACKGROUND: Diabetic cardiomyopathy (DCM) is a leading cause of death in diabetic patients. Hyperglycemic myocardial microenvironment significantly alters chromatin architecture and the transcriptome, resulting in aberrant activation of signaling pathways in a diabetic heart. Epigenetic marks play vital roles in transcriptional reprogramming during the development of DCM. The current study is aimed to profile genome-wide DNA (hydroxy)methylation patterns in the hearts of control and streptozotocin (STZ)-induced diabetic rats and decipher the effect of modulation of DNA methylation by alpha-ketoglutarate (AKG), a TET enzyme cofactor, on the progression of DCM. METHODS: Diabetes was induced in male adult Wistar rats with an intraperitoneal injection of STZ. Diabetic and vehicle control animals were randomly divided into groups with/without AKG treatment. Cardiac function was monitored by performing cardiac catheterization. Global methylation (5mC) and hydroxymethylation (5hmC) patterns were mapped in the Left ventricular tissue of control and diabetic rats with the help of an enrichment-based (h)MEDIP-sequencing technique by using antibodies specific for 5mC and 5hmC. Sequencing data were validated by performing (h)MEDIP-qPCR analysis at the gene-specific level, and gene expression was analyzed by qPCR. The mRNA and protein expression of enzymes involved in the DNA methylation and demethylation cycle were analyzed by qPCR and western blotting. Global 5mC and 5hmC levels were also assessed in high glucose-treated DNMT3B knockdown H9c2 cells. RESULTS: We found the increased expression of DNMT3B, MBD2, and MeCP2 with a concomitant accumulation of 5mC and 5hmC, specifically in gene body regions of diabetic rat hearts compared to the control. Calcium signaling was the most significantly affected pathway by cytosine modifications in the diabetic heart. Additionally, hypermethylated gene body regions were associated with Rap1, apelin, and phosphatidyl inositol signaling, while metabolic pathways were most affected by hyperhydroxymethylation. AKG supplementation in diabetic rats reversed aberrant methylation patterns and restored cardiac function. Hyperglycemia also increased 5mC and 5hmC levels in H9c2 cells, which was normalized by DNMT3B knockdown or AKG supplementation. CONCLUSION: This study demonstrates that reverting hyperglycemic damage to cardiac tissue might be possible by erasing adverse epigenetic signatures by supplementing epigenetic modulators such as AKG along with an existing antidiabetic treatment regimen.</p>',
'date' => '2023-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37101286',
'doi' => '10.1186/s13072-023-00489-4',
'modified' => '2023-06-12 09:20:54',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '4780',
'name' => 'Integrated analysis from multicentre studies identities RNAmethylation- related lncRNA risk stratification systems for glioma',
'authors' => 'Huang Fanxuan and Wang Xinyu and Zhong Junzhe and Chen Hao and Song Dan and Xu Tianye and Tian Kaifu and Sun Penggang and Sun Nan and Ma Wenbin and Liu Yuxiang andYu Daohan and Meng Xiangqi and Jiang Chuanlu and Xuan Hanwen and Qian Da an',
'description' => '<p>Gastric cancer (GC) is the fourth leading cause of cancer death worldwide. Due to the lack of effective chemotherapy methods for advanced gastric cancer and poor prognosis, the emergence of immunotherapy has brought new hope to gastric cancer. Further research is needed to improve the response rate to immunotherapy and identify the populations with potential benefits of immunotherapy. It is unclear whether m7G-related lncRNAs influence tumour immunity and the prognosis of immunotherapy.</p>',
'date' => '2023-03-02',
'pmid' => 'https://doi.org/10.21203%2Frs.3.rs-2815231%2Fv1',
'doi' => '10.21203/rs.3.rs-2815231/v1',
'modified' => '2023-06-13 09:25:12',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '4773',
'name' => 'The RNA m5C Methylase NSUN2 Modulates Corneal EpithelialWound Healing.',
'authors' => 'Luo G. et al.',
'description' => '<p>PURPOSE: The emerging epitranscriptomics offers insights into the physiopathological roles of various RNA modifications. The RNA methylase NOP2/Sun domain family member 2 (NSUN2) catalyzes 5-methylcytosine (m5C) modification of mRNAs. However, the role of NSUN2 in corneal epithelial wound healing (CEWH) remains unknown. Here we describe the functional mechanisms of NSUN2 in mediating CEWH. METHODS: RT-qPCR, Western blot, dot blot, and ELISA were used to determine the NSUN2 expression and overall RNA m5C level during CEWH. NSUN2 silencing or overexpression was performed to explore its involvement in CEWH both in vivo and in vitro. Multi-omics was integrated to reveal the downstream target of NSUN2. MeRIP-qPCR, RIP-qPCR, and luciferase assay, as well as in vivo and in vitro functional assays, clarified the molecular mechanism of NSUN2 in CEWH. RESULTS: The NSUN2 expression and RNA m5C level increased significantly during CEWH. NSUN2 knockdown significantly delayed CEWH in vivo and inhibited human corneal epithelial cells (HCEC) proliferation and migration in vitro, whereas NSUN2 overexpression prominently enhanced HCEC proliferation and migration. Mechanistically, we found that NSUN2 increased ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) translation through the binding of RNA m5C reader Aly/REF export factor. Accordingly, UHRF1 knockdown significantly delayed CEWH in vivo and inhibited HCEC proliferation and migration in vitro. Furthermore, UHRF1 overexpression effectively rescued the inhibitory effect of NSUN2 silencing on HCEC proliferation and migration. CONCLUSIONS: NSUN2-mediated m5C modification of UHRF1 mRNA modulates CEWH. This finding highlights the critical importance of this novel epitranscriptomic mechanism in control of CEWH.</p>',
'date' => '2023-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36862118',
'doi' => '10.1167/iovs.64.3.5',
'modified' => '2023-04-17 09:48:55',
'created' => '2023-04-14 13:41:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '4674',
'name' => 'Methylation and expression of glucocorticoid receptor exon-1 variants andFKBP5 in teenage suicide-completers.',
'authors' => 'Rizavi H. et al.',
'description' => '<p>A dysregulated hypothalamic-pituitary-adrenal (HPA) axis has repeatedly been demonstrated to play a fundamental role in psychiatric disorders and suicide, yet the mechanisms underlying this dysregulation are not clear. Decreased expression of the glucocorticoid receptor (GR) gene, which is also susceptible to epigenetic modulation, is a strong indicator of impaired HPA axis control. In the context of teenage suicide-completers, we have systematically analyzed the 5'UTR of the GR gene to determine the expression levels of all GR exon-1 transcript variants and their epigenetic state. We also measured the expression and the epigenetic state of the FK506-binding protein 51 (FKBP5/FKBP51), an important modulator of GR activity. Furthermore, steady-state DNA methylation levels depend upon the interplay between enzymes that promote DNA methylation and demethylation activities, thus we analyzed DNA methyltransferases (DNMTs), ten-eleven translocation enzymes (TETs), and growth arrest- and DNA-damage-inducible proteins (GADD45). Focusing on both the prefrontal cortex (PFC) and hippocampus, our results show decreased expression in specific GR exon-1 variants and a strong correlation of DNA methylation changes with gene expression in the PFC. FKBP5 expression is also increased in both areas suggesting a decreased GR sensitivity to cortisol binding. We also identified aberrant expression of DNA methylating and demethylating enzymes in both brain regions. These findings enhance our understanding of the complex transcriptional regulation of GR, providing evidence of epigenetically mediated reprogramming of the GR gene, which could lead to possible epigenetic influences that result in lasting modifications underlying an individual's overall HPA axis response and resilience to stress.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36781843',
'doi' => '10.1038/s41398-023-02345-1',
'modified' => '2023-04-14 09:26:37',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '4675',
'name' => 'Bridging biological cfDNA features and machine learning approaches.',
'authors' => 'Moser T. et al.',
'description' => '<p>Liquid biopsies (LBs), particularly using circulating tumor DNA (ctDNA), are expected to revolutionize precision oncology and blood-based cancer screening. Recent technological improvements, in combination with the ever-growing understanding of cell-free DNA (cfDNA) biology, are enabling the detection of tumor-specific changes with extremely high resolution and new analysis concepts beyond genetic alterations, including methylomics, fragmentomics, and nucleosomics. The interrogation of a large number of markers and the high complexity of data render traditional correlation methods insufficient. In this regard, machine learning (ML) algorithms are increasingly being used to decipher disease- and tissue-specific signals from cfDNA. Here, we review recent insights into biological ctDNA features and how these are incorporated into sophisticated ML applications.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36792446',
'doi' => '10.1016/j.tig.2023.01.004',
'modified' => '2023-04-14 09:28:00',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '4823',
'name' => 'Gene body DNA hydroxymethylation restricts the magnitude oftranscriptional changes during aging.',
'authors' => 'Occean J. R. et al.',
'description' => '<p>DNA hydroxymethylation (5hmC) is the most abundant oxidative derivative of DNA methylation (5mC) and is typically enriched at enhancers and gene bodies of transcriptionally active and tissue-specific genes. Although aberrant genomic 5hmC has been implicated in many age-related diseases, the functional role of the modification in aging remains largely unknown. Here, we report that 5hmC is stably enriched in multiple aged organs. Using the liver and cerebellum as model organs, we show that 5hmC accumulates in gene bodies associated with tissue-specific function and thereby restricts the magnitude of gene expression changes during aging. Mechanistically, we found that 5hmC decreases binding affinity of splicing factors compared to unmodified cytosine and 5mC, and is correlated with age-related alternative splicing events, suggesting RNA splicing as a potential mediator of 5hmC’s transcriptionally restrictive function. Furthermore, we show that various age-related contexts, such as prolonged quiescence and senescence, are partially responsible for driving the accumulation of 5hmC with age. We provide evidence that this age-related function is conserved in mouse and human tissues, and further show that the modification is altered by regimens known to modulate lifespan. Our findings reveal that 5hmC is a regulator of tissue-specific function and may play a role in regulating longevity.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36824863',
'doi' => '10.1101/2023.02.15.528714',
'modified' => '2023-06-14 08:39:26',
'created' => '2023-06-13 22:16:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '4631',
'name' => 'Consistent DNA Hypomethylations in Prostate Cancer.',
'authors' => 'Araúzo-Bravo M.J. et al.',
'description' => '<p>With approximately 1.4 million men annually diagnosed with prostate cancer (PCa) worldwide, PCa remains a dreaded threat to life and source of devastating morbidity. In recent decades, a significant decrease in age-specific PCa mortality has been achieved by increasing prostate-specific antigen (PSA) screening and improving treatments. Nevertheless, upcoming, augmented recommendations against PSA screening underline an escalating disproportion between the benefit and harm of current diagnosis/prognosis and application of radical treatment standards. Undoubtedly, new potent diagnostic and prognostic tools are urgently needed to alleviate this tensed situation. They should allow a more reliable early assessment of the upcoming threat, in order to enable applying timely adjusted and personalized therapy and monitoring. Here, we present a basic study on an epigenetic screening approach by Methylated DNA Immunoprecipitation (MeDIP). We identified genes associated with hypomethylated CpG islands in three PCa sample cohorts. By adjusting our computational biology analyses to focus on single CpG-enriched 60-nucleotide-long DNA probes, we revealed numerous consistently differential methylated DNA segments in PCa. They were associated among other genes with and . These can be used for early discrimination, and might contribute to a new epigenetic tumor classification system of PCa. Our analysis shows that we can dissect short, differential methylated CpG-rich DNA fragments and combinations of them that are consistently present in all tumors. We name them tumor cell-specific differential methylated CpG dinucleotide signatures (TUMS).</p>',
'date' => '2022-12-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36613831',
'doi' => '10.3390/ijms24010386',
'modified' => '2023-03-28 09:03:47',
'created' => '2023-02-21 09:59:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '4534',
'name' => 'RNA 5-Methylcytosine Modification Regulates VegetativeDevelopment Associated with H3K27 Trimethylation inArabidopsis.',
'authors' => 'Zhang D.et al.',
'description' => '<p>Methylating RNA post-transcriptionally is emerging as a significant mechanism of gene regulation in eukaryotes. The crosstalk between RNA methylation and histone modification is critical for chromatin state and gene expression in mammals. However, it is not well understood mechanistically in plants. Here, the authors report a genome-wide correlation between RNA 5-cytosine methylation (m C) and histone 3 lysine27 trimethylation (H3K27me3) in Arabidopsis. The plant-specific Polycomb group (PcG) protein EMBRYONIC FLOWER1 (EMF1) plays dual roles as activators or repressors. Transcriptome-wide RNA m C profiling revealed that m C peaks are mostly enriched in chromatin regions that lacked H3K27me3 in both wild type and emf1 mutants. EMF1 repressed the expression of m C methyltransferase tRNA specific methyltransferase 4B (TRM4B) through H3K4me3, independent of PcG-mediated H3K27me3 mechanism. The 5-Cytosine methylation on targets is increased in emf1 mutants, thereby decreased the mRNA transcripts of photosynthesis and chloroplast genes. In addition, impairing EMF1 activity reduced H3K27me3 levels of PcG targets, such as starch genes, which are de-repressed in emf1 mutants. Both EMF1-mediated promotion and repression of gene activities via m C and H3K27me3 are required for normal vegetative growth. Collectively, t study reveals a previously undescribed epigenetic mechanism of RNA m C modifications and histone modifications to regulate gene expression in eukaryotes.</p>',
'date' => '2022-11-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36382558',
'doi' => '10.1002/advs.202204885',
'modified' => '2022-11-24 08:57:31',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '4541',
'name' => 'Cardiac epigenetic changes in VEGF signaling genes associates with myocardial microvascular rarefaction in experimental chronic kidney disease.',
'authors' => 'Eirin Alfonso and Chade Alejandro R',
'description' => '<p>BACKGROUND: Chronic kidney disease (CKD) is common in patients with heart failure, and often results in left ventricular diastolic dysfunction (LVDD). However, the mechanisms responsible for cardiac damage in CKD-LVDD remain to be elucidated. Epigenetic alterations may impose long-lasting effects on cellular transcription and function, but their exact role in CKD-LVDD is unknown. We investigate whether changes in cardiac site-specific DNA methylation profiles might be implicated in cardiac abnormalities in CKD-LVDD. METHODS: CKD-LVDD and normal control pigs (n=6 each) were studied for 14 weeks. Renal and cardiac hemodynamics were quantified by multidetector CT and echocardiography. In randomly selected pigs (n=3/group), cardiac site-specific 5-methylcytosine (5mC) immunoprecipitation (MeDIP)- and mRNA-sequencing (seq) was performed, followed by integrated (MeDiP-seq/mRNA-seq analysis), and confirmatory ex vivo studies. RESULTS: MeDIP-seq analysis revealed 261 genes with higher (fold-change>1.4; p<0.05) and 162 genes with lower (fold-change<0.7; p<0.05) 5mC levels in CKD-LVDD versus normal pigs, which were primarily implicated in vascular endothelial growth factor (VEGF)-related signaling and angiogenesis. Integrated MeDiP-seq/mRNA-seq analysis identified a select group of VEGF-related genes in which 5mC levels were higher, but mRNA expression lower in CKD-LVDD versus normal pigs. Cardiac VEGF signaling gene and VEGF protein expression was blunted in CKD-LVDD compared to controls and associated with decreased subendocardial microvascular density. CONCLUSIONS: Cardiac epigenetic changes in VEGF-related genes are associated with impaired angiogenesis and cardiac microvascular rarefaction in swine CKD-LVDD. These observations may assist in developing novel therapies to ameliorate cardiac damage in CKD-LVDD.</p>',
'date' => '2022-11-01',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/36367693/',
'doi' => '10.1152/ajpheart.00522.2022',
'modified' => '2022-11-25 09:03:31',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '4511',
'name' => 'The Arabidopsis APOLO and human UPAT sequence-unrelated longnoncoding RNAs can modulate DNA and histone methylation machineries inplants.',
'authors' => 'Fonouni-Farde C. et al.',
'description' => '<p>BACKGROUND: RNA-DNA hybrid (R-loop)-associated long noncoding RNAs (lncRNAs), including the Arabidopsis lncRNA AUXIN-REGULATED PROMOTER LOOP (APOLO), are emerging as important regulators of three-dimensional chromatin conformation and gene transcriptional activity. RESULTS: Here, we show that in addition to the PRC1-component LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), APOLO interacts with the methylcytosine-binding protein VARIANT IN METHYLATION 1 (VIM1), a conserved homolog of the mammalian DNA methylation regulator UBIQUITIN-LIKE CONTAINING PHD AND RING FINGER DOMAINS 1 (UHRF1). The APOLO-VIM1-LHP1 complex directly regulates the transcription of the auxin biosynthesis gene YUCCA2 by dynamically determining DNA methylation and H3K27me3 deposition over its promoter during the plant thermomorphogenic response. Strikingly, we demonstrate that the lncRNA UHRF1 Protein Associated Transcript (UPAT), a direct interactor of UHRF1 in humans, can be recognized by VIM1 and LHP1 in plant cells, despite the lack of sequence homology between UPAT and APOLO. In addition, we show that increased levels of APOLO or UPAT hamper VIM1 and LHP1 binding to YUCCA2 promoter and globally alter the Arabidopsis transcriptome in a similar manner. CONCLUSIONS: Collectively, our results uncover a new mechanism in which a plant lncRNA coordinates Polycomb action and DNA methylation through the interaction with VIM1, and indicates that evolutionary unrelated lncRNAs with potentially conserved structures may exert similar functions by interacting with homolog partners.</p>',
'date' => '2022-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36038910',
'doi' => '10.1186/s13059-022-02750-7',
'modified' => '2022-11-21 10:43:16',
'created' => '2022-11-15 09:26:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '4438',
'name' => 'A genome-wide screen reveals new regulators of the 2-cell-like cell state',
'authors' => 'Defossez Pierre-Antoine et al.',
'description' => '<p>In mammals, only the zygote and blastomeres of the early embryo are fully totipotent. This totipotency is mirrored in vitro by mouse "2-cell-like cells" (2CLCs), which appear at low frequency in cultures of Embryonic Stem cells (ESCs). Because totipotency is incompletely understood, we carried out a genomewide CRISPR KO screen in mouse ESCs, searching for mutants that reactivate the expression of Dazl, a robust 2-cell-like marker. Using secondary screens, we identify four mutants that reactivate not just Dazl, but also a broader 2-cell-like signature: the E3 ubiquitin ligase adaptor SPOP, the Zinc Finger transcription factor ZBTB14, MCM3AP, a component of the RNA processing complex TREX-2, and the lysine demethylase KDM5C. Functional experiments show how these factors link to known players of the 2 celllike state. These results extend our knowledge of totipotency, a key phase of organismal life.</p>',
'date' => '2022-06-01',
'pmid' => 'https://doi.org/10.21203%2Frs.3.rs-1561018%2Fv1',
'doi' => '10.21203/rs.3.rs-1561018/v1',
'modified' => '2022-09-28 09:23:42',
'created' => '2022-09-08 16:32:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '4553',
'name' => 'NSUN2-mediated RNA mC modification modulates uveal melanoma cellproliferation and migration.',
'authors' => 'Luo Guangying et al.',
'description' => '<p>RNA 5-methylcytosine (mC) is a widespread post-transcriptional modification involved in diverse biological processes through controlling RNA metabolism. However, its roles in uveal melanoma (UM) remain unknown. Here, we describe the biological roles and regulatory mechanisms of RNA mC in UM. Initially, we identified significantly elevated global RNA mC levels in both UM cells and tissue specimens using ELISA assay and dot blot analysis. Meanwhile, NOP2/Sun RNA methyltransferase family member 2 (NSUN2) was upregulated in both types of these samples, whereas NSUN2 knockdown significantly decreased RNA mC level. Such declines inhibited UM cell migration and suppressed cell proliferation through cell cycle G1 arrest. Furthermore, bioinformatic analyses, mC-RIP-qPCR, and luciferase assay identified β-Catenin (CTNNB1) as a direct target of NSUN2-mediated mC modification in UM cells. Additionally, overexpression of miR-124a in UM cells diminished NSUN2 expression levels indicating that it is an upstream regulator of this response. Our study suggests that NSUN2-mediated RNA mC methylation provides a potential novel target to improve the therapeutic management of UM pathogenesis.</p>',
'date' => '2022-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/35757999',
'doi' => '10.1080/15592294.2022.2088047',
'modified' => '2022-11-24 10:14:24',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '4340',
'name' => 'Global DNA methylation and cellular 5-methylcytosine and H4acetylated patterns in primary and secondary dormant seeds of Capsellabursa-pastoris (L.) Medik. (shepherd's purse).',
'authors' => 'Gomez-Cabellos Sara et al.',
'description' => '<p>Despite the importance of dormancy and dormancy cycling for plants' fitness and life cycle phenology, a comprehensive characterization of the global and cellular epigenetic patterns across space and time in different seed dormancy states is lacking. Using Capsella bursa-pastoris (L.) Medik. (shepherd's purse) seeds with primary and secondary dormancy, we investigated the dynamics of global genomic DNA methylation and explored the spatio-temporal distribution of 5-methylcytosine (5-mC) and histone H4 acetylated (H4Ac) epigenetic marks. Seeds were imbibed at 30 °C in a light regime to maintain primary dormancy, or in darkness to induce secondary dormancy. An ELISA-based method was used to quantify DNA methylation, in relation to total genomic cytosines. Immunolocalization of 5-mC and H4Ac within whole seeds (i.e., including testa) was assessed with reference to embryo anatomy. Global DNA methylation levels were highest in prolonged (14 days) imbibed primary dormant seeds, with more 5-mC marked nuclei present only in specific parts of the seed (e.g., SAM and cotyledons). In secondary dormant seeds, global methylation levels and 5-mC signal where higher at 3 and 7 days than 1 or 14 days. With respect to acetylation, seeds had fewer H4Ac marked nuclei (e.g., SAM) in deeper dormant states, for both types of dormancy. However, the RAM still showed signal after 14 days of imbibition under dormancy-inducing conditions, suggesting a central role for the radicle/RAM in the response to perceived ambient changes and the adjustment of the seed dormancy state. Thus, we show that seed dormancy involves extensive cellular remodeling of DNA methylation and H4 acetylation.</p>',
'date' => '2022-05-01',
'pmid' => 'https://doi.org/10.1007%2Fs00709-021-01678-2',
'doi' => '10.1007/s00709-021-01678-2',
'modified' => '2022-06-20 09:19:49',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '4404',
'name' => 'Stella regulates the Development of Female Germline Stem Cells byModulating Chromatin Structure and DNA Methylation.',
'authors' => 'Hou Changliang et al.',
'description' => '<p>Female germline stem cells (FGSCs) have the ability to self-renew and differentiate into oocytes. , encoded by a maternal effect gene, plays an important role in oogenesis and early embryonic development. However, its function in FGSCs remains unclear. In this study, we showed that CRISPR/Cas9-mediated knockout of promoted FGSC proliferation and reduced the level of genome-wide DNA methylation of FGSCs. Conversely, overexpression led to the opposite results, and enhanced FGSC differentiation. We also performed an integrative analysis of chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), high-throughput genome-wide chromosome conformation capture (Hi-C), and use of our published epigenetic data. Results indicated that the binding sites of STELLA and active histones H3K4me3 and H3K27ac were enriched near the TAD boundaries. Hi-C analysis showed that overexpression attenuated the interaction within TADs, and interestingly enhanced the TAD boundary strength in STELLA-associated regions. Taking these findings together, our study not only reveals the role of in regulating DNA methylation and chromatin structure, but also provides a better understanding of FGSC development.</p>',
'date' => '2022-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9066111/',
'doi' => '10.7150/ijbs.69240',
'modified' => '2022-08-11 14:54:29',
'created' => '2022-08-11 12:14:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '4327',
'name' => 'Highly recurrent epimutations in gastric cancer CpG islandmethylator phenotypes and inflammation',
'authors' => 'Padmanabhan N. et al.',
'description' => '<p>Background CIMP (CpG island methylator phenotype) is an epigenetic molecular subtype, observed in multiple malignancies and associated with the epigenetic silencing of tumor suppressors. Currently, for most cancers including gastric cancer (GC), mechanisms underlying CIMP remain poorly understood. We sought to discover molecular contributors to CIMP in GC, by performing global DNA methylation, gene expression, and proteomics profiling across 14 gastric cell lines, followed by similar integrative analysis in 50 GC cell lines and 467 primary GCs. Results We identify the cystathionine beta-synthase enzyme (CBS) as a highly recurrent target of epigenetic silencing in CIMP GC. Likewise, we show that CBS epimutations are significantly associated with CIMP in various other cancers, occurring even in premalignant gastroesophageal conditions and longitudinally linked to clinical persistence. Of note, CRISPR deletion of CBS in normal gastric epithelial cells induces widespread DNA methylation changes that overlap with primary GC CIMP patterns. Reflecting its metabolic role as a gatekeeper interlinking the methionine and homocysteine cycles, CBS loss in vitro also causes reductions in the anti-inflammatory gasotransmitter hydrogen sulfide (H2S), with concomitant increase in NF-κB activity. In a murine genetic model of CBS deficiency, preliminary data indicate upregulated immune-mediated transcriptional signatures in the stomach. Conclusions Our results implicate CBS as a bi-faceted modifier of aberrant DNA methylation and inflammation in GC and highlights H2S donors as a potential new therapy for CBS-silenced lesions. Supplementary Information The online version contains supplementary material available at 10.1186/s13059-021-02375-2.</p>',
'date' => '2021-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/34074348',
'doi' => '10.1186/s13059-021-02375-2',
'modified' => '2022-08-03 16:01:40',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '4336',
'name' => 'LINE-1 transcription in round spermatids is associated with accretion of5-carboxylcytosine in their open reading frames',
'authors' => 'Blythe M. et al. ',
'description' => '<p>Chromatin of male and female gametes undergoes a number of reprogramming events during the transition from germ cell to embryonic developmental programs. Although the rearrangement of DNA methylation patterns occurring in the zygote has been extensively characterized, little is known about the dynamics of DNA modifications during spermatid maturation. Here, we demonstrate that the dynamics of 5-carboxylcytosine (5caC) correlate with active transcription of LINE-1 retroelements during murine spermiogenesis. We show that the open reading frames of active and evolutionary young LINE-1s are 5caC-enriched in round spermatids and 5caC is eliminated from LINE-1s and spermiogenesis-specific genes during spermatid maturation, being simultaneously retained at promoters and introns of developmental genes. Our results reveal an association of 5caC with activity of LINE-1 retrotransposons suggesting a potential direct role for this DNA modification in fine regulation of their transcription.</p>',
'date' => '2021-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/34099857',
'doi' => '10.1038/s42003-021-02217-8',
'modified' => '2022-08-03 16:17:04',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '4150',
'name' => 'Sensitive and reproducible cell-free methylome quantification with synthetic spike-in controls',
'authors' => 'Wilson, S.L. et al.',
'description' => '<p>Background. Cell-free methylated DNA immunoprecipitation-sequencing (cfMeDIP-seq) identifies genomic regions with DNA methylation, using a protocol adapted to work with low-input DNA samples and with cell-free DNA (cfDNA). This method allows for DNA methylation profiling of circulating tumour DNA in cancer patients’ blood samples. Such epigenetic profiling of circulating tumour DNA provides information about in which tissues tumour DNA originates, a key requirement of any test for early cancer detection. In addition, DNA methylation signatures provide prognostic information and can detect relapse. For robust quantitative comparisons between samples, immunoprecipitation enrichment methods like cfMeDIP-seq require normalization against common reference controls.</p>',
'date' => '2021-04-01',
'pmid' => 'https://doi.org/10.1101%2F2021.02.12.430289',
'doi' => '10.1101/2021.02.12.430289',
'modified' => '2022-01-13 15:16:23',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '3985',
'name' => 'Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes.',
'authors' => 'Nuzzo PV, Berchuck JE, Korthauer K, Spisak S, Nassar AH, Abou Alaiwi S, Chakravarthy A, Shen SY, Bakouny Z, Boccardo F, Steinharter J, Bouchard G, Curran CR, Pan W, Baca SC, Seo JH, Lee GM, Michaelson MD, Chang SL, Waikar SS, Sonpavde G, Irizarry RA, Pome',
'description' => '<p>Improving early cancer detection has the potential to substantially reduce cancer-related mortality. Cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) is a highly sensitive assay capable of detecting early-stage tumors. We report accurate classification of patients across all stages of renal cell carcinoma (RCC) in plasma (area under the receiver operating characteristic (AUROC) curve of 0.99) and demonstrate the validity of this assay to identify patients with RCC using urine cell-free DNA (cfDNA; AUROC of 0.86).</p>',
'date' => '2020-06-22',
'pmid' => 'http://www.pubmed.gov/32572266',
'doi' => '10.1038/s41591-020-0933-1',
'modified' => '2020-09-01 15:13:49',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '3984',
'name' => 'Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes.',
'authors' => 'Nassiri F, Chakravarthy A, Feng S, Shen SY, Nejad R, Zuccato JA, Voisin MR, Patil V, Horbinski C, Aldape K, Zadeh G, De Carvalho DD',
'description' => '<p>Definitive diagnosis of intracranial tumors relies on tissue specimens obtained by invasive surgery. Noninvasive diagnostic approaches provide an opportunity to avoid surgery and mitigate unnecessary risk to patients. In the present study, we show that DNA-methylation profiles from plasma reveal highly specific signatures to detect and accurately discriminate common primary intracranial tumors that share cell-of-origin lineages and can be challenging to distinguish using standard-of-care imaging.</p>',
'date' => '2020-06-22',
'pmid' => 'http://www.pubmed.gov/32572265',
'doi' => '10.1038/s41591-020-0932-2',
'modified' => '2020-09-01 15:14:45',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '4030',
'name' => 'AXR1 affects DNA methylation independently of its role in regulatingmeiotic crossover localization.',
'authors' => 'Christophorou, N and She, W and Long, J and Hurel, A and Beaubiat, S andIdir, Y and Tagliaro-Jahns, M and Chambon, A and Solier, V and Vezon, D andGrelon, M and Feng, X and Bouché, N and Mézard, C',
'description' => '<p>Meiotic crossovers (COs) are important for reshuffling genetic information between homologous chromosomes and they are essential for their correct segregation. COs are unevenly distributed along chromosomes and the underlying mechanisms controlling CO localization are not well understood. We previously showed that meiotic COs are mis-localized in the absence of AXR1, an enzyme involved in the neddylation/rubylation protein modification pathway in Arabidopsis thaliana. Here, we report that in axr1-/-, male meiocytes show a strong defect in chromosome pairing whereas the formation of the telomere bouquet is not affected. COs are also redistributed towards subtelomeric chromosomal ends where they frequently form clusters, in contrast to large central regions depleted in recombination. The CO suppressed regions correlate with DNA hypermethylation of transposable elements (TEs) in the CHH context in axr1-/- meiocytes. Through examining somatic methylomes, we found axr1-/- affects DNA methylation in a plant, causing hypermethylation in all sequence contexts (CG, CHG and CHH) in TEs. Impairment of the main pathways involved in DNA methylation is epistatic over axr1-/- for DNA methylation in somatic cells but does not restore regular chromosome segregation during meiosis. Collectively, our findings reveal that the neddylation pathway not only regulates hormonal perception and CO distribution but is also, directly or indirectly, a major limiting pathway of TE DNA methylation in somatic cells.</p>',
'date' => '2020-06-01',
'pmid' => 'http://www.pubmed.gov/32598340',
'doi' => '10.1371/journal.',
'modified' => '2020-12-16 17:58:51',
'created' => '2020-10-12 14:54:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '3951',
'name' => 'In vitro capture and characterization of embryonic rosette-stage pluripotency between naive and primed states.',
'authors' => 'Neagu A, van Genderen E, Escudero I, Verwegen L, Kurek D, Lehmann J, Stel J, Dirks RAM, van Mierlo G, Maas A, Eleveld C, Ge Y, den Dekker AT, Brouwer RWW, van IJcken WFJ, Modic M, Drukker M, Jansen JH, Rivron NC, Baart EB, Marks H, Ten Berge D',
'description' => '<p>Following implantation, the naive pluripotent epiblast of the mouse blastocyst generates a rosette, undergoes lumenogenesis and forms the primed pluripotent egg cylinder, which is able to generate the embryonic tissues. How pluripotency progression and morphogenesis are linked and whether intermediate pluripotent states exist remain controversial. We identify here a rosette pluripotent state defined by the co-expression of naive factors with the transcription factor OTX2. Downregulation of blastocyst WNT signals drives the transition into rosette pluripotency by inducing OTX2. The rosette then activates MEK signals that induce lumenogenesis and drive progression to primed pluripotency. Consequently, combined WNT and MEK inhibition supports rosette-like stem cells, a self-renewing naive-primed intermediate. Rosette-like stem cells erase constitutive heterochromatin marks and display a primed chromatin landscape, with bivalently marked primed pluripotency genes. Nonetheless, WNT induces reversion to naive pluripotency. The rosette is therefore a reversible pluripotent intermediate whereby control over both pluripotency progression and morphogenesis pivots from WNT to MEK signals.</p>',
'date' => '2020-05-01',
'pmid' => 'http://www.pubmed.gov/32367046',
'doi' => '10.1038/s41556-020-0508-x',
'modified' => '2020-08-17 09:55:37',
'created' => '2020-08-10 12:12:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '3834',
'name' => 'Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.',
'authors' => 'Liu J, An Z, Luo J, Li J, Li F, Zhang Z',
'description' => '<p>MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of transcriptome-wide profile and potential function of m5C in splicing remains to be elucidated due to lack of isoform level m5C quantification tool. RESULTS: We developed a computational package to quantify Epitranscriptomal RNA m5C at the transcript isoform level (named Episo). Episo consists of three tools, mapper, quant and Bisulfitefq, for mapping, quantifying, and simulating RNA-BisSeq data, respectively. The high accuracy of Episo was validated using an improved m5C-specific methylated RNA immunoprecipitation (meRIP) protocol, as well as a set of in silico experiments. By applying Episo to public human and mouse RNA-BisSeq data, we found that the RNA m5C is not evenly distributed among the transcript isoforms, implying the m5C may subject to be regulated at isoform level. AVAILABILITY: Episo is released under the GNU GPLv3+ license. The resource code Episo is freely accessible from https://github.com/liujunfengtop/Episo (with Tophat/cufflink) and https://github.com/liujunfengtop/Episo/tree/master/Episo_Kallisto (with Kallisto). SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.</p>',
'date' => '2019-12-03',
'pmid' => 'http://www.pubmed.gov/31794005',
'doi' => '10.1093/bioinformatics/btz900/5651015',
'modified' => '2020-02-25 13:26:22',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 25 => array(
'id' => '3827',
'name' => 'Intra- and inter-generational changes in the cortical DNA methylome in response to therapeutic intermittent hypoxia in mice.',
'authors' => 'Belmonte KCD, Harman JC, Lanson NA, Gidday JM',
'description' => '<p>Recent evidence from our lab documents functional resilience to retinal ischemic injury in untreated mice derived from parents exposed to repetitive hypoxic conditioning (RHC) prior to breeding. To begin to understand the epigenetic basis of this intergenerational protection, we used methylated DNA immunoprecipitation and sequencing to identify genes with differentially-methylated promoters (DMGPs) in the prefrontal cortex of mice treated directly with the same RHC stimulus (F0-RHC), and in the prefrontal cortex of their untreated F1-generation offspring (F1-*RHC). Subsequent bioinformatic analyses provided key mechanistic insights into how changes in gene expression secondary to promoter hypo- and hyper-methylation might afford such protection within and across generations. We found extensive changes in DNA methylation in both generations consistent with the expression of many survival-promoting genes, with twice the number of DMGPs in the cortex of F1*RHC mice relative to their F0 parents that were directly exposed to RHC. In contrast to our hypothesis that similar epigenetic modifications would be realized in the cortices of both F0-RHC and F1-*RHC mice, we instead found relatively few DMGPs common to both generations; in fact, each generation manifested expected injury resilience via distinctly unique gene expression profiles. Whereas in the cortex of F0-RHC mice, predicted protein-protein interactions reflected the activation of an anti-ischemic phenotype, networks activated in F1-*RHC cortex comprised networks indicative of a much broader cytoprotective phenotype. Altogether, our results suggest that the intergenerational transfer of an acquired phenotype to offspring does not necessarily require the faithful recapitulation of the conditioning-modified DNA methylome of the parent.</p>',
'date' => '2019-11-25',
'pmid' => 'http://www.pubmed.gov/31762411',
'doi' => '10.1152/physiolgenomics.00094.2019',
'modified' => '2020-02-25 13:35:09',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 26 => array(
'id' => '3814',
'name' => 'Lithium treatment reverses irradiation-induced changes in rodent neural progenitors and rescues cognition.',
'authors' => 'Zanni G, Goto S, Fragopoulou AF, Gaudenzi G, Naidoo V, Di Martino E, Levy G, Dominguez CA, Dethlefsen O, Cedazo-Minguez A, Merino-Serrais P, Stamatakis A, Hermanson O, Blomgren K',
'description' => '<p>Cranial radiotherapy in children has detrimental effects on cognition, mood, and social competence in young cancer survivors. Treatments harnessing hippocampal neurogenesis are currently of great relevance in this context. Lithium, a well-known mood stabilizer, has both neuroprotective, pro-neurogenic as well as antitumor effects, and in the current study we introduced lithium treatment 4 weeks after irradiation. Female mice received a single 4 Gy whole-brain radiation dose on postnatal day (PND) 21 and were randomized to 0.24% Li2CO chow or normal chow from PND 49 to 77. Hippocampal neurogenesis was assessed on PND 77, 91, and 105. We found that lithium treatment had a pro-proliferative effect on neural progenitors, but neuronal integration occurred only after it was discontinued. Also, the treatment ameliorated deficits in spatial learning and memory retention observed in irradiated mice. Gene expression profiling and DNA methylation analysis identified two novel factors related to the observed effects, Tppp, associated with microtubule stabilization, and GAD2/65, associated with neuronal signaling. Our results show that lithium treatment reverses irradiation-induced loss of hippocampal neurogenesis and cognitive impairment even when introduced long after the injury. We propose that lithium treatment should be intermittent in order to first make neural progenitors proliferate and then, upon discontinuation, allow them to differentiate. Our findings suggest that pharmacological treatment of cognitive so-called late effects in childhood cancer survivors is possible.</p>',
'date' => '2019-11-14',
'pmid' => 'http://www.pubmed.gov/31723242',
'doi' => '10.1038/s41380-019-0584-0',
'modified' => '2019-12-05 10:58:44',
'created' => '2019-12-02 15:25:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 27 => array(
'id' => '3773',
'name' => 'Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA.',
'authors' => 'Shen SY, Burgener JM, Bratman SV, De Carvalho DD',
'description' => '<p>Circulating cell-free DNA (cfDNA) comprises small DNA fragments derived from normal and tumor tissue that are released into the bloodstream. Recently, methylation profiling of cfDNA as a liquid biopsy tool has been gaining prominence due to the presence of tissue-specific markers in cfDNA. We have previously reported cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) as a sensitive, low-input, cost-efficient and bisulfite-free approach to profiling DNA methylomes of plasma cfDNA. cfMeDIP-seq is an extension of a previously published MeDIP-seq protocol and is adapted to allow for methylome profiling of samples with low input (ranging from 1 to 10 ng) of DNA, which is enabled by the addition of 'filler DNA' before immunoprecipitation. This protocol is not limited to plasma cfDNA; it can also be applied to other samples that are naturally sheared and at low availability (e.g., urinary cfDNA and cerebrospinal fluid cfDNA), and is potentially applicable to other applications beyond cancer detection, including prenatal diagnostics, cardiology and monitoring of immune response. The protocol presented here should enable any standard molecular laboratory to generate cfMeDIP-seq libraries from plasma cfDNA in ~3-4 d.</p>',
'date' => '2019-08-30',
'pmid' => 'http://www.pubmed.gov/31471598',
'doi' => '10.1038/s41596-019-0202-2',
'modified' => '2019-10-02 17:07:45',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 28 => array(
'id' => '3763',
'name' => 'Silencing of tumor-suppressive NR_023387 in renal cell carcinoma via promoter hypermethylation and HNF4A deficiency.',
'authors' => 'Zhou H, Guo L, Yao W, Shi R, Yu G, Xu H, Ye Z',
'description' => '<p>Dysregulation of the epigenetic status of long noncoding RNAs (lncRNAs) has been linked to diverse human diseases including human cancers. However, the landscape of the whole-genome methylation profile of lncRNAs and the precise roles of these lncRNAs remain elusive in renal cell carcinoma (RCC). We first examined lncRNA expression profiles in RCC tissues and corresponding adjacent normal tissues (NTs) to identify the lncRNA signature of RCC, then lncRNA Promoter Microarray was performed to depict the whole-genome methylation profile of lncRNAs in RCC. Combined analysis of the lncRNAs expression profiles and lncRNAs Promoter Microarray identified a series of downregulated lncRNAs with hypermethylated promoter regions, including NR_023387. Quantitative real-time polymerase chain reaction (RT-PCR) implied that NR_023387 was significantly downregulated in RCC tissues and cell lines, and lower expression of NR_023387 was correlated with shorter overall survival. Methylation-specific PCR, MassARRAY, and demethylation drug treatment indicated that hypermethylation in the NR_023387 promoter contributed to its silencing in RCC. Besides, HNF4A regulated the expression of NR_023387 via transcriptional activation. Functional experiments demonstrated NR_023387 exerted tumor-suppressive roles in RCC via suppressing the proliferation, migration, invasion, tumor growth, and metastasis of RCC. Furthermore, we identified MGP as a putative downstream molecule of NR_023387, which promoted the epithelial-mesenchymal transition of RCC cells. Our study provides the first whole-genome lncRNA methylation profile in RCC. Our combined analysis identifies a tumor-suppressive and prognosis-related lncRNA NR_023387, which is silenced in RCC via promoter hypermethylation and HNF4A deficiency, and may exert its tumor-suppressive roles by downregulating the oncogenic MGP.</p>',
'date' => '2019-08-20',
'pmid' => 'http://www.pubmed.gov/31432508',
'doi' => '10.1002/jcp.29115',
'modified' => '2019-10-03 10:02:27',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 29 => array(
'id' => '3770',
'name' => 'Epitranscriptomic Addition of mC to HIV-1 Transcripts Regulates Viral Gene Expression.',
'authors' => 'Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR',
'description' => '<p>How the covalent modification of mRNA ribonucleotides, termed epitranscriptomic modifications, alters mRNA function remains unclear. One issue has been the difficulty of quantifying these modifications. Using purified HIV-1 genomic RNA, we show that this RNA bears more epitranscriptomic modifications than the average cellular mRNA, with 5-methylcytosine (mC) and 2'O-methyl modifications being particularly prevalent. The methyltransferase NSUN2 serves as the primary writer for mC on HIV-1 RNAs. NSUN2 inactivation inhibits not only mC addition to HIV-1 transcripts but also viral replication. This inhibition results from reduced HIV-1 protein, but not mRNA, expression, which in turn correlates with reduced ribosome binding to viral mRNAs. In addition, loss of mC dysregulates the alternative splicing of viral RNAs. These data identify mC as a post-transcriptional regulator of both splicing and function of HIV-1 mRNA, thereby affecting directly viral gene expression.</p>',
'date' => '2019-08-14',
'pmid' => 'http://www.pubmed.gov/31415754',
'doi' => '10.1016/j.chom.2019.07.005',
'modified' => '2019-10-03 09:18:50',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 30 => array(
'id' => '3741',
'name' => 'Aberrant expression of imprinted lncRNA MEG8 causes trophoblast dysfunction and abortion.',
'authors' => 'Sheng F, Sun N, Ji Y, Ma Y, Ding H, Zhang Q, Yang F, Li W',
'description' => '<p>Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs whose nucleotides are longer than 200 bp. Previous studies have shown that they play an important regulatory role in many developmental processes and biological pathways. However, the contributions of lncRNAs to placental development are largely unknown. Here, our study aimed to investigate the lncRNA expression signatures in placental development by performing a microarray lncRNA screen. Placental samples were obtained from pregnant C57BL/6 female mice at three key developmental time points (embryonic day E7.5, E13.5, and E19.5). Microarrays were used to analyze the differential expression of lncRNAs during placental development. In addition to the genomic imprinting region and the dynamic DNA methylation status during placental development, we screened imprinted lncRNAs whose expression was controlled by DNA methylation during placental development. We found that the imprinted lncRNA Rian may play an important role during placental development. Its homologous sequence lncRNA MEG8 (RIAN) was abnormally highly expressed in human spontaneous abortion villi. Upregulation of MEG8 expression in trophoblast cell lines decreased cell proliferation and invasion, whereas downregulation of MEG8 expression had the opposite effect. Furthermore, DNA methylation results showed that the methylation of the MEG8 promoter region was increased in spontaneous abortion villi. There was dynamic spatiotemporal expression of imprinted lncRNAs during placental development. The imprinted lncRNA MEG8 is involved in the regulation of early trophoblast cell function. Promoter methylation abnormalities can cause trophoblastic cell defects, which may be one of the factors that occurs in early unexplained spontaneous abortion.</p>',
'date' => '2019-07-02',
'pmid' => 'http://www.pubmed.gov/31265183',
'doi' => '10.1002/jcb.29002',
'modified' => '2019-08-06 16:45:53',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 31 => array(
'id' => '3731',
'name' => 'Defining UHRF1 Domains that Support Maintenance of Human Colon Cancer DNA Methylation and Oncogenic Properties.',
'authors' => 'Kong X, Chen J, Xie W, Brown SM, Cai Y, Wu K, Fan D, Nie Y, Yegnasubramanian S, Tiedemann RL, Tao Y, Chiu Yen RW, Topper MJ, Zahnow CA, Easwaran H, Rothbart SB, Xia L, Baylin SB',
'description' => '<p>UHRF1 facilitates the establishment and maintenance of DNA methylation patterns in mammalian cells. The establishment domains are defined, including E3 ligase function, but the maintenance domains are poorly characterized. Here, we demonstrate that UHRF1 histone- and hemimethylated DNA binding functions, but not E3 ligase activity, maintain cancer-specific DNA methylation in human colorectal cancer (CRC) cells. Disrupting either chromatin reader activity reverses DNA hypermethylation, reactivates epigenetically silenced tumor suppressor genes (TSGs), and reduces CRC oncogenic properties. Moreover, an inverse correlation between high UHRF1 and low TSG expression tracks with CRC progression and reduced patient survival. Defining critical UHRF1 domain functions and its relationship with CRC prognosis suggests directions for, and value of, targeting this protein to develop therapeutic DNA demethylating agents.</p>',
'date' => '2019-04-15',
'pmid' => 'http://www.pubmed.gov/30956060',
'doi' => '10.1016/j.ccell.2019.03.003',
'modified' => '2019-08-07 09:14:54',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 32 => array(
'id' => '3729',
'name' => 'Tricarboxylic Acid Cycle Activity and Remodeling of Glycerophosphocholine Lipids Support Cytokine Induction in Response to Fungal Patterns.',
'authors' => 'Márquez S, Fernández JJ, Mancebo C, Herrero-Sánchez C, Alonso S, Sandoval TA, Rodríguez Prados M, Cubillos-Ruiz JR, Montero O, Fernández N, Sánchez Crespo M',
'description' => '<p>Increased glycolysis parallels immune cell activation, but the role of pyruvate remains largely unexplored. We found that stimulation of dendritic cells with the fungal surrogate zymosan causes decreases of pyruvate, citrate, itaconate, and α-ketoglutarate, while increasing oxaloacetate, succinate, lactate, oxygen consumption, and pyruvate dehydrogenase activity. Expression of IL10 and IL23A (the gene encoding the p19 chain of IL-23) depended on pyruvate dehydrogenase activity. Mechanistically, pyruvate reinforced histone H3 acetylation, and acetate rescued the effect of mitochondrial pyruvate carrier inhibition, most likely because it is a substrate of the acetyl-CoA producing enzyme ACSS2. Mice lacking the receptor of the lipid mediator platelet-activating factor (PAF; 1-O-hexadecyl-2-acetyl-sn-glycero-3-phosphocholine) showed reduced production of IL-10 and IL-23 that is explained by the requirement of acetyl-CoA for PAF biosynthesis and its ensuing autocrine function. Acetyl-CoA therefore intertwines fatty acid remodeling of glycerophospholipids and energetic metabolism during cytokine induction.</p>',
'date' => '2019-04-09',
'pmid' => 'http://www.pubmed.gov/30970255',
'doi' => '10.1016/j.celrep.2019.03.033',
'modified' => '2019-08-07 09:15:46',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 33 => array(
'id' => '3693',
'name' => 'Increased Serine and One-Carbon Pathway Metabolism by PKCλ/ι Deficiency Promotes Neuroendocrine Prostate Cancer.',
'authors' => 'Reina-Campos M, Linares JF, Duran A, Cordes T, L'Hermitte A, Badur MG, Bhangoo MS, Thorson PK, Richards A, Rooslid T, Garcia-Olmo DC, Nam-Cha SY, Salinas-Sanchez AS, Eng K, Beltran H, Scott DA, Metallo CM, Moscat J, Diaz-Meco MT',
'description' => '<p>Increasingly effective therapies targeting the androgen receptor have paradoxically promoted the incidence of neuroendocrine prostate cancer (NEPC), the most lethal subtype of castration-resistant prostate cancer (PCa), for which there is no effective therapy. Here we report that protein kinase C (PKC)λ/ι is downregulated in de novo and during therapy-induced NEPC, which results in the upregulation of serine biosynthesis through an mTORC1/ATF4-driven pathway. This metabolic reprogramming supports cell proliferation and increases intracellular S-adenosyl methionine (SAM) levels to feed epigenetic changes that favor the development of NEPC characteristics. Altogether, we have uncovered a metabolic vulnerability triggered by PKCλ/ι deficiency in NEPC, which offers potentially actionable targets to prevent therapy resistance in PCa.</p>',
'date' => '2019-03-18',
'pmid' => 'http://www.pubmed.gov/30827887',
'doi' => '10.1016/j.ccell.2019.01.018',
'modified' => '2019-06-28 13:49:24',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 34 => array(
'id' => '3660',
'name' => 'Global distribution of DNA hydroxymethylation and DNA methylation in chronic lymphocytic leukemia.',
'authors' => 'Wernig-Zorc S, Yadav MP, Kopparapu PK, Bemark M, Kristjansdottir HL, Andersson PO, Kanduri C, Kanduri M',
'description' => '<p>BACKGROUND: Chronic lymphocytic leukemia (CLL) has been a good model system to understand the functional role of 5-methylcytosine (5-mC) in cancer progression. More recently, an oxidized form of 5-mC, 5-hydroxymethylcytosine (5-hmC) has gained lot of attention as a regulatory epigenetic modification with prognostic and diagnostic implications for several cancers. However, there is no global study exploring the role of 5-hydroxymethylcytosine (5-hmC) levels in CLL. Herein, using mass spectrometry and hMeDIP-sequencing, we analysed the dynamics of 5-hmC during B cell maturation and CLL pathogenesis. RESULTS: We show that naïve B-cells had higher levels of 5-hmC and 5-mC compared to non-class switched and class-switched memory B-cells. We found a significant decrease in global 5-mC levels in CLL patients (n = 15) compared to naïve and memory B cells, with no changes detected between the CLL prognostic groups. On the other hand, global 5-hmC levels of CLL patients were similar to memory B cells and reduced compared to naïve B cells. Interestingly, 5-hmC levels were increased at regulatory regions such as gene-body, CpG island shores and shelves and 5-hmC distribution over the gene-body positively correlated with degree of transcriptional activity. Importantly, CLL samples showed aberrant 5-hmC and 5-mC pattern over gene-body compared to well-defined patterns in normal B-cells. Integrated analysis of 5-hmC and RNA-sequencing from CLL datasets identified three novel oncogenic drivers that could have potential roles in CLL development and progression. CONCLUSIONS: Thus, our study suggests that the global loss of 5-hmC, accompanied by its significant increase at the gene regulatory regions, constitute a novel hallmark of CLL pathogenesis. Our combined analysis of 5-mC and 5-hmC sequencing provided insights into the potential role of 5-hmC in modulating gene expression changes during CLL pathogenesis.</p>',
'date' => '2019-01-07',
'pmid' => 'http://www.pubmed.gov/30616658',
'doi' => '10.1186/s13072‑018‑0252‑7',
'modified' => '2019-07-01 11:46:16',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 35 => array(
'id' => '3730',
'name' => 'Transcriptome-Wide Mapping 5-Methylcytosine by mC RNA Immunoprecipitation Followed by Deep Sequencing in Plant.',
'authors' => 'Gu X, Liang Z',
'description' => '<p>Transcriptome-wide mapping RNA modification is crucial to understand the distribution and function of RNA modifications. Here, we describe a protocol to transcriptome-wide mapping 5-methylcytosine (mC) in plant, by a RNA immunoprecipitation followed by deep sequencing (mC-RIP-seq) approach. The procedure includes RNA extraction, fragmentation, RNA immunoprecipitation, and library construction.</p>',
'date' => '2019-01-01',
'pmid' => 'http://www.pubmed.gov/30945199',
'doi' => '10.1007/978-1-4939-9045-0_24,',
'modified' => '2019-08-07 10:21:37',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 36 => array(
'id' => '3584',
'name' => 'Sensitivity of pituitary gonadotropes to hyperglycemia leads to epigenetic aberrations and reduced follicle-stimulating hormone levels.',
'authors' => 'Feldman A, Saleh A, Pnueli L, Qiao S, Shlomi T, Boehm U, Melamed P',
'description' => '<p>The connection between metabolism and reproductive function is well recognized, and we hypothesized that the pituitary gonadotropes, which produce luteinizing hormone and follicle-stimulating hormone (FSH), mediate some of the effects directly via insulin-independent glucose transporters, which allow continued glucose metabolism during hyperglycemia. We found that glucose transporter 1 is the predominant glucose transporter in primary gonadotropes and a gonadotrope precursor-derived cell line, and both are responsive to culture in high glucose; moreover, metabolite levels were altered in the cell line. Several of the affected metabolites are cofactors for chromatin-modifying enzymes, and in the gonadotrope precursor-derived cell line, we recorded global changes in histone acetylation and methylation, decreased DNA methylation, and increased hydroxymethylation, some of which did not revert to basal levels after cells were returned to normal glucose. Despite this weakening of epigenetic-mediated repression seen in the model cell line, FSH β-subunit ( Fshb) mRNA levels in primary gonadotropes were significantly reduced, apparently due in part to increased autocrine/paracrine effects of inhibin. However, unlike thioredoxin interacting protein and inhibin subunit α, Fshb mRNA levels did not recover after the return of cells to normal glucose. The effect on Fshb expression was also seen in 2 hyperglycemic mouse models, and levels of circulating FSH, required for follicle growth and development, were reduced. Thus, hyperglycemia seems to target the pituitary gonadotropes directly, and the likely extensive epigenetic changes are sensed acutely by Fshb. This scenario would explain clinical findings in which, even after restoration of optimal blood glucose levels, fertility often remains adversely affected. However, the relative accessibility of the pituitary provides a possible target for treatment, particularly crucial in the young in which hyperglycemia is increasingly common and fertility most relevant.-Feldman, A., Saleh, A., Pnueli, L., Qiao, S., Shlomi, T., Boehm, U., Melamed, P. Sensitivity of pituitary gonadotropes to hyperglycemia leads to epigenetic aberrations and reduced follicle-stimulating hormone levels.</p>',
'date' => '2018-12-27',
'pmid' => 'http://www.pubmed.gov/30074825',
'doi' => '10.1096/fj.201800943R',
'modified' => '2019-04-17 15:48:51',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 37 => array(
'id' => '3430',
'name' => 'Sensitive tumour detection and classification using plasma cell-free DNA methylomes.',
'authors' => 'Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, Kis O, Zhao Z, Spreafico A, Medina TDS, Wang Y, Roulois D, Ettayebi I, Chen Z, Chow S, Murphy T, Arruda A, O'Kane GM, Liu J, Mansour M, McPher',
'description' => '<p>The use of liquid biopsies for cancer detection and management is rapidly gaining prominence. Current methods for the detection of circulating tumour DNA involve sequencing somatic mutations using cell-free DNA, but the sensitivity of these methods may be low among patients with early-stage cancer given the limited number of recurrent mutations. By contrast, large-scale epigenetic alterations-which are tissue- and cancer-type specific-are not similarly constrained and therefore potentially have greater ability to detect and classify cancers in patients with early-stage disease. Here we develop a sensitive, immunoprecipitation-based protocol to analyse the methylome of small quantities of circulating cell-free DNA, and demonstrate the ability to detect large-scale DNA methylation changes that are enriched for tumour-specific patterns. We also demonstrate robust performance in cancer detection and classification across an extensive collection of plasma samples from several tumour types. This work sets the stage to establish biomarkers for the minimally invasive detection, interception and classification of early-stage cancers based on plasma cell-free DNA methylation patterns.</p>',
'date' => '2018-11-14',
'pmid' => 'http://www.pubmed.gov/30429608',
'doi' => '10.1038/s41586-018-0703-0',
'modified' => '2019-06-11 16:22:54',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 38 => array(
'id' => '3421',
'name' => 'Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate.',
'authors' => 'Sardina JL, Collombet S, Tian TV, Gómez A, Di Stefano B, Berenguer C, Brumbaugh J, Stadhouders R, Segura-Morales C, Gut M, Gut IG, Heath S, Aranda S, Di Croce L, Hochedlinger K, Thieffry D, Graf T',
'description' => '<p>Here, we report DNA methylation and hydroxymethylation dynamics at nucleotide resolution using C/EBPα-enhanced reprogramming of B cells into induced pluripotent cells (iPSCs). We observed successive waves of hydroxymethylation at enhancers, concomitant with a decrease in DNA methylation, suggesting active demethylation. Consistent with this finding, ablation of the DNA demethylase Tet2 almost completely abolishes reprogramming. C/EBPα, Klf4, and Tfcp2l1 each interact with Tet2 and recruit the enzyme to specific DNA sites. During reprogramming, some of these sites maintain high levels of 5hmC, and enhancers and promoters of key pluripotency factors become demethylated as early as 1 day after Yamanaka factor induction. Surprisingly, methylation changes precede chromatin opening in distinct chromatin regions, including Klf4 bound sites, revealing a pioneer factor activity associated with alternation in DNA methylation. Rapid changes in hydroxymethylation similar to those in B cells were also observed during compound-accelerated reprogramming of fibroblasts into iPSCs, highlighting the generality of our observations.</p>',
'date' => '2018-11-01',
'pmid' => 'http://www.pubmed.gov/30220521',
'doi' => '10.1016/j.stem.2018.08.016',
'modified' => '2018-12-31 11:16:24',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 39 => array(
'id' => '3409',
'name' => 'Oxidative stress in sperm affects the epigenetic reprogramming in early embryonic development.',
'authors' => 'Wyck S, Herrera C, Requena CE, Bittner L, Hajkova P, Bollwein H, Santoro R',
'description' => '<p>BACKGROUND: Reactive oxygen species (ROS)-induced oxidative stress is well known to play a major role in male infertility. Sperm are sensitive to ROS damaging effects because as male germ cells form mature sperm they progressively lose the ability to repair DNA damage. However, how oxidative DNA lesions in sperm affect early embryonic development remains elusive. RESULTS: Using cattle as model, we show that fertilization using sperm exposed to oxidative stress caused a major developmental arrest at the time of embryonic genome activation. The levels of DNA damage response did not directly correlate with the degree of developmental defects. The early cellular response for DNA damage, γH2AX, is already present at high levels in zygotes that progress normally in development and did not significantly increase at the paternal genome containing oxidative DNA lesions. Moreover, XRCC1, a factor implicated in the last step of base excision repair (BER) pathway, was recruited to the damaged paternal genome, indicating that the maternal BER machinery can repair these DNA lesions induced in sperm. Remarkably, the paternal genome with oxidative DNA lesions showed an impairment of zygotic active DNA demethylation, a process that previous studies linked to BER. Quantitative immunofluorescence analysis and ultrasensitive LC-MS-based measurements revealed that oxidative DNA lesions in sperm impair active DNA demethylation at paternal pronuclei, without affecting 5-hydroxymethylcytosine (5hmC), a 5-methylcytosine modification that has been implicated in paternal active DNA demethylation in mouse zygotes. Thus, other 5hmC-independent processes are implicated in active DNA demethylation in bovine embryos. The recruitment of XRCC1 to damaged paternal pronuclei indicates that oxidative DNA lesions drive BER to repair DNA at the expense of DNA demethylation. Finally, this study highlighted striking differences in DNA methylation dynamics between bovine and mouse zygotes that will facilitate the understanding of the dynamics of DNA methylation in early development. CONCLUSIONS: The data demonstrate that oxidative stress in sperm has an impact not only on DNA integrity but also on the dynamics of epigenetic reprogramming, which may harm the paternal genetic and epigenetic contribution to the developing embryo and affect embryo development and embryo quality.</p>',
'date' => '2018-10-17',
'pmid' => 'http://www.pubmed.gov/30333056',
'doi' => '10.1186/s13072-018-0224-y',
'modified' => '2018-11-09 11:10:58',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 40 => array(
'id' => '3404',
'name' => 'Integrated analysis of DNA methylation profiling and gene expression profiling identifies novel markers in lung cancer in Xuanwei, China.',
'authors' => 'Wang J, Duan Y, Meng QH, Gong R, Guo C, Zhao Y, Zhang Y',
'description' => '<p>BACKGROUND: Aberrant DNA methylation occurs frequently in cancer. The aim of this study was to identify novel methylation markers in lung cancer in Xuanwei, China, through integrated genome-wide DNA methylation and gene expression studies. METHODS: Differentially methylated regions (DMRs) and differentially expressed genes (DEGs) were detected on 10 paired lung cancer tissues and noncancerous lung tissues by methylated DNA immunoprecipitation combined with microarray (MeDIP-chip) and gene expression microarray analyses, respectively. Integrated analysis of DMRs and DEGs was performed to screen out candidate methylation-related genes. Both methylation and expression changes of the candidate genes were further validated and analyzed. RESULTS: Compared with normal lung tissues, lung cancer tissues expressed a total of 6,899 DMRs, including 5,788 hypermethylated regions and 1,111 hypomethylated regions. Integrated analysis of DMRs and DEGs identified 45 tumor-specific candidate genes: 38 genes whose DMRs were hypermethylated and expression was downregulated, and 7 genes whose DMRs were hypomethylated and expression was upregulated. The methylation and expression validation results identified 4 candidate genes (STXBP6, BCL6B, FZD10, and HSPB6) that were significantly hypermethylated and downregulated in most of the tumor tissues compared with the noncancerous lung tissues. CONCLUSIONS: This integrated analysis of genome-wide DNA methylation and gene expression in lung cancer in Xuanwei revealed several genes regulated by promoter methylation that have not been described in lung cancer before. These results provide new insight into the carcinogenesis of lung cancer in Xuanwei and represent promising new diagnostic and therapeutic targets.</p>',
'date' => '2018-10-04',
'pmid' => 'http://www.pubmed.gov/30286088',
'doi' => '10.1371/journal.pone.0203155',
'modified' => '2018-11-09 11:24:21',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 41 => array(
'id' => '3417',
'name' => 'mGlu1 Receptors Monopolize the Synaptic Control of Cerebellar Purkinje Cells by Epigenetically Down-Regulating mGlu5 Receptors.',
'authors' => 'Notartomaso S, Nakao H, Mascio G, Scarselli P, Cannella M, Zappulla C, Madonna M, Motolese M, Gradini R, Liberatore F, Zonta M, Carmignoto G, Battaglia G, Bruno V, Watanabe M, Aiba A, Nicoletti F',
'description' => '<p>In cerebellar Purkinje cells (PCs) type-1 metabotropic glutamate (mGlu1) receptors play a key role in motor learning and drive the refinement of synaptic innervation during postnatal development. The cognate mGlu5 receptor is absent in mature PCs and shows low expression levels in the adult cerebellar cortex. Here we found that mGlu5 receptors were heavily expressed by PCs in the early postnatal life, when mGlu1α receptors were barely detectable. The developmental decline of mGlu5 receptors coincided with the appearance of mGlu1α receptors in PCs, and both processes were associated with specular changes in CpG methylation in the corresponding gene promoters. It was the mGlu1 receptor that drove the elimination of mGlu5 receptors from PCs, as shown by data obtained with conditional mGlu1α receptor knockout mice and with targeted pharmacological treatments during critical developmental time windows. The suppressing activity of mGlu1 receptors on mGlu5 receptor was maintained in mature PCs, suggesting that expression of mGlu1α and mGlu5 receptors is mutually exclusive in PCs. These findings add complexity to the the finely tuned mechanisms that regulate PC biology during development and in the adult life and lay the groundwork for an in-depth analysis of the role played by mGlu5 receptors in PC maturation.</p>',
'date' => '2018-09-06',
'pmid' => 'http://www.pubmed.gov/30190524',
'doi' => '10.1038/s41598-018-31369-7',
'modified' => '2018-12-31 11:36:04',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 42 => array(
'id' => '3640',
'name' => 'Determination of the presence of 5-methylcytosine in Paramecium tetraurelia.',
'authors' => 'Singh A, Vancura A, Woycicki RK, Hogan DJ, Hendrick AG, Nowacki M',
'description' => '<p>5-methylcytosine DNA methylation regulates gene expression and developmental programming in a broad range of eukaryotes. However, its presence and potential roles in ciliates, complex single-celled eukaryotes with germline-somatic genome specialization via nuclear dimorphism, are largely uncharted. While canonical cytosine methyltransferases have not been discovered in published ciliate genomes, recent studies performed in the stichotrichous ciliate Oxytricha trifallax suggest de novo cytosine methylation during macronuclear development. In this study, we applied bisulfite genome sequencing, DNA mass spectrometry and antibody-based fluorescence detection to investigate the presence of DNA methylation in Paramecium tetraurelia. While the antibody-based methods suggest cytosine methylation, DNA mass spectrometry and bisulfite sequencing reveal that levels are actually below the limit of detection. Our results suggest that Paramecium does not utilize 5-methylcytosine DNA methylation as an integral part of its epigenetic arsenal.</p>',
'date' => '2018-06-07',
'pmid' => 'http://www.pubmed.gov/30379964',
'doi' => '10.1371/journal.',
'modified' => '2019-06-07 10:22:28',
'created' => '2019-06-06 12:11:18',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 43 => array(
'id' => '3458',
'name' => 'Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.',
'authors' => 'Li T, Wang L, Du Y, Xie S, Yang X, Lian F, Zhou Z, Qian C',
'description' => '<p>UHRF1 plays multiple roles in regulating DNMT1-mediated DNA methylation maintenance during DNA replication. The UHRF1 C-terminal RING finger functions as an ubiquitin E3 ligase to establish histone H3 ubiquitination at Lys18 and/or Lys23, which is subsequently recognized by DNMT1 to promote its localization onto replication foci. Here, we present the crystal structure of DNMT1 RFTS domain in complex with ubiquitin and highlight a unique ubiquitin binding mode for the RFTS domain. We provide evidence that UHRF1 N-terminal ubiquitin-like domain (UBL) also binds directly to DNMT1. Despite sharing a high degree of structural similarity, UHRF1 UBL and ubiquitin bind to DNMT1 in a very distinct fashion and exert different impacts on DNMT1 enzymatic activity. We further show that the UHRF1 UBL-mediated interaction between UHRF1 and DNMT1, and the binding of DNMT1 to ubiquitinated histone H3 that is catalyzed by UHRF1 RING domain are critical for the proper subnuclear localization of DNMT1 and maintenance of DNA methylation. Collectively, our study adds another layer of complexity to the regulatory mechanism of DNMT1 activation by UHRF1 and supports that individual domains of UHRF1 participate and act in concert to maintain DNA methylation patterns.</p>',
'date' => '2018-04-06',
'pmid' => 'http://www.pubmed.gov/29471350',
'doi' => '10.1093/nar/gky104',
'modified' => '2019-02-15 21:14:42',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 44 => array(
'id' => '3475',
'name' => 'Epigenetics and early domestication: differences in hypothalamic DNA methylation between red junglefowl divergently selected for high or low fear of humans.',
'authors' => 'Bélteky J, Agnvall B, Bektic L, Höglund A, Jensen P, Guerrero-Bosagna C',
'description' => '<p>BACKGROUND: Domestication of animals leads to large phenotypic alterations within a short evolutionary time-period. Such alterations are caused by genomic variations, yet the prevalence of modified traits is higher than expected if they were caused only by classical genetics and mutations. Epigenetic mechanisms may also be important in driving domesticated phenotypes such as behavior traits. Gene expression can be modulated epigenetically by mechanisms such as DNA methylation, resulting in modifications that are not only variable and susceptible to environmental stimuli, but also sometimes transgenerationally stable. To study such mechanisms in early domestication, we used as model two selected lines of red junglefowl (ancestors of modern chickens) that were bred for either high or low fear of humans over five generations, and investigated differences in hypothalamic DNA methylation between the two populations. RESULTS: Twenty-two 1-kb windows were differentially methylated between the two selected lines at p < 0.05 after false discovery rate correction. The annotated functions of the genes within these windows indicated epigenetic regulation of metabolic and signaling pathways, which agrees with the changes in gene expression that were previously reported for the same tissue and animals. CONCLUSIONS: Our results show that selection for an important domestication-related behavioral trait such as tameness can cause divergent epigenetic patterns within only five generations, and that these changes could have an important role in chicken domestication.</p>',
'date' => '2018-04-02',
'pmid' => 'http://www.pubmed.gov/29609558',
'doi' => '10.1186/s12711-018-0384-z',
'modified' => '2019-02-15 20:32:37',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 45 => array(
'id' => '3393',
'name' => 'Copper induces expression and methylation changes of early development genes in Crassostrea gigas embryos.',
'authors' => 'Sussarellu R, Lebreton M, Rouxel J, Akcha F, Rivière G',
'description' => '<p>Copper contamination is widespread along coastal areas and exerts adverse effects on marine organisms such as mollusks. In the Pacific oyster, copper induces severe developmental abnormalities during early life stages; however, the underlying molecular mechanisms are largely unknown. This study aims to better understand whether the embryotoxic effects of copper in Crassostrea gigas could be mediated by alterations in gene expression, and the putative role of DNA methylation, which is known to contribute to gene regulation in early embryo development. For that purpose, oyster embryos were exposed to 4 nominal copper concentrations (0.1, 1, 10 and 20 μg L Cu) during early development assays. Embryotoxicity was monitored through the oyster embryo-larval bioassay at the D-larva stage 24 h post fertilization (hpf) and genotoxicity at gastrulation 7 hpf. In parallel, the relative expression of 15 genes encoding putative homeotic, biomineralization and DNA methylation proteins was measured at three developmental stages (3 hpf morula stage, 7 hpf gastrula stage, 24 hpf D-larvae stage) using RT-qPCR. Global DNA content in methylcytosine and hydroxymethylcytosine were measured by HPLC and gene-specific DNA methylation levels were monitored using MeDIP-qPCR. A significant increase in larval abnormalities was observed from copper concentrations of 10 μg L, while significant genotoxic effects were detected at 1 μg L and above. All the selected genes presented a stage-dependent expression pattern, which was impaired for some homeobox and DNA methylation genes (Notochord, HOXA1, HOX2, Lox5, DNMT3b and CXXC-1) after copper exposure. While global DNA methylation (5-methylcytosine) at gastrula stage didn't show significant changes between experimental conditions, 5-hydroxymethylcytosine, its degradation product, decreased upon copper treatment. The DNA methylation of exons and the transcript levels were correlated in control samples for HOXA1 but such a correlation was diminished following copper exposure. The methylation level of some specific gene regions (HoxA1, Hox2, Engrailed2 and Notochord) displayed changes upon copper exposure. Such changes were gene and exon-specific and no obvious global trends could be identified. Our study suggests that the embryotoxic effects of copper in oysters could involve homeotic gene expression impairment possibly by changing DNA methylation levels.</p>',
'date' => '2018-03-01',
'pmid' => 'http://www.pubmed.gov/29353135',
'doi' => '10.1016/j.aquatox.2018.01.001',
'modified' => '2018-11-09 12:21:38',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 46 => array(
'id' => '3448',
'name' => 'Aberrant methylated key genes of methyl group metabolism within the molecular etiology of urothelial carcinogenesis.',
'authors' => 'Erichsen L, Ghanjati F, Beermann A, Poyet C, Hermanns T, Schulz WA, Seifert HH, Wild PJ, Buser L, Kröning A, Braunstein S, Anlauf M, Jankowiak S, Hassan M, Bendhack ML, Araúzo-Bravo MJ, Santourlidis S',
'description' => '<p>Urothelial carcinoma (UC), the most common cancer of the urinary bladder causes severe morbidity and mortality, e.g. about 40.000 deaths in the EU annually, and incurs considerable costs for the health system due to the need for prolonged treatments and long-term monitoring. Extensive aberrant DNA methylation is described to prevail in urothelial carcinoma and is thought to contribute to genetic instability, altered gene expression and tumor progression. However, it is unknown how this epigenetic alteration arises during carcinogenesis. Intact methyl group metabolism is required to ensure maintenance of cell-type specific methylomes and thereby genetic integrity and proper cellular function. Here, using two independent techniques for detecting DNA methylation, we observed DNA hypermethylation of the 5'-regulatory regions of the key methyl group metabolism genes ODC1, AHCY and MTHFR in early urothelial carcinoma. These hypermethylation events are associated with genome-wide DNA hypomethylation which is commonly associated with genetic instability. We therefore infer that hypermethylation of methyl group metabolism genes acts in a feed-forward cycle to promote additional DNA methylation changes and suggest a new hypothesis on the molecular etiology of urothelial carcinoma.</p>',
'date' => '2018-02-22',
'pmid' => 'http://www.pubmed.gov/29472622',
'doi' => '10.1038/s41598-018-21932-7',
'modified' => '2019-02-15 21:31:04',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 47 => array(
'id' => '3383',
'name' => 'Genome-wide analysis of day/night DNA methylation differences in Populus nigra.',
'authors' => 'Ding C.J. et al.',
'description' => '<p>DNA methylation is an important mechanism of epigenetic modification. Methylation changes during stress responses and developmental processes have been well studied; however, their role in plant adaptation to the day/night cycle is poorly understood. In this study, we detected global methylation patterns in leaves of the black poplar Populus nigra 'N46' at 8:00 and 24:00 by methylated DNA immunoprecipitation sequencing (MeDIP-seq). We found 10,027 and 10,242 genes to be methylated in the 8:00 and 24:00 samples, respectively. The methylated genes appeared to be involved in multiple biological processes, molecular functions, and cellular components, suggesting important roles for DNA methylation in poplar cells. Comparing the 8:00 and 24:00 samples, only 440 differentially methylated regions (DMRs) overlapped with genic regions, including 193 hyper- and 247 hypo-methylated DMRs, and may influence the expression of 137 downstream genes. Most hyper-methylated genes were associated with transferase activity, kinase activity, and phosphotransferase activity, whereas most hypo-methylated genes were associated with protein binding, ATP binding, and adenyl ribonucleotide binding, suggesting that different biological processes were activated during the day and night. Our results indicated that methylated genes were prevalent in the poplar genome, but that only a few of these participated in diurnal gene expression regulation.</p>',
'date' => '2018-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29293569',
'doi' => '',
'modified' => '2018-08-07 09:45:38',
'created' => '2018-08-07 09:45:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 48 => array(
'id' => '3384',
'name' => 'Obligatory and facilitative allelic variation in the DNA methylome within common disease-associated loci',
'authors' => 'Bell C.G. et al.',
'description' => '<p>Integrating epigenetic data with genome-wide association study (GWAS) results can reveal disease mechanisms. The genome sequence itself also shapes the epigenome, with CpG density and transcription factor binding sites (TFBSs) strongly encoding the DNA methylome. Therefore, genetic polymorphism impacts on the observed epigenome. Furthermore, large genetic variants alter epigenetic signal dosage. Here, we identify DNA methylation variability between GWAS-SNP risk and non-risk haplotypes. In three subsets comprising 3128 MeDIP-seq peripheral-blood DNA methylomes, we find 7173 consistent and functionally enriched Differentially Methylated Regions. 36.8% can be attributed to common non-SNP genetic variants. CpG-SNPs, as well as facilitative TFBS-motifs, are also enriched. Highlighting their functional potential, CpG-SNPs strongly associate with allele-specific DNase-I hypersensitivity sites. Our results demonstrate strong DNA methylation allelic differences driven by obligatory or facilitative genetic effects, with potential direct or regional disease-related repercussions. These allelic variations require disentangling from pure tissue-specific modifications, may influence array studies, and imply underestimated population variability in current reference epigenomes.</p>',
'date' => '2018-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29295990',
'doi' => '',
'modified' => '2018-08-07 10:13:12',
'created' => '2018-08-07 10:13:12',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 49 => array(
'id' => '3508',
'name' => 'Analysis of DNA methylome and transcriptome profiling following Gibberellin A3 (GA3) foliar application in Nicotiana tabacum L.',
'authors' => 'Manoharlal Raman, Saiprasad G. V. S., Kaikala Vinay, Suresh Kumar R., Kovařík Ales',
'description' => '<p>The present work investigated a comprehensive genome-wide landscape of DNA methylome and its relationship with transcriptome upon gibberellin A3 (GA3) foliar application under practical field conditions in solanaceae model, Nicotiana tabacum L. Methylated DNA Immunoprecipitation-Sequencing (MeDIP-Seq) analysis uncovered over 82% (18,456) of differential methylated regions (DMRs) in intergenic-region. Within protein-coding region, 2339 and 1685 of identified DMRs were observed in genebody- and promoter-region, respectively. Microarray study revealed 7032 differential expressed genes (DEGs) with 3507 and 3525 genes displaying upand down-regulation, respectively. Integration analysis revealed 520 unique non-redundant annotated DMRs overlapping with DEGs. Our results indicated that GA3 induced DNA hypo- as well as hyper-methylation were associated with both gene-silencing and -activation. No complete biasness or correlation was observed in either of the promoter- or genebody-regions, which otherwise showed an overall trend towards GA3 induced global DNA hypo-methylation. Taken together, our results suggested that differential DNA methylation mediated by GA3 may only play a permissive role in regulating the gene expression.</p>',
'date' => '2018-01-01',
'pmid' => 'https://link.springer.com/article/10.1007/s40502-018-0393-5',
'doi' => '10.1007/s40502-018-0393-5',
'modified' => '2022-05-18 18:43:47',
'created' => '2019-02-27 12:54:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 50 => array(
'id' => '3297',
'name' => '5-Methylcytosine RNA Methylation in Arabidopsis Thaliana',
'authors' => 'Cui X. et al.',
'description' => '<p>5-Methylcytosine (m<sup>5</sup>C) is a well-characterized DNA modification, and is also predominantly reported in abundant non-coding RNAs in both prokaryotes and eukaryotes. However, the distribution and biological functions of m<sup>5</sup>C in plant mRNAs remain largely unknown. Here, we report transcriptome-wide profiling of RNA m<sup>5</sup>C in Arabidopsis thaliana by applying m<sup>5</sup>C RNA immunoprecipitation followed by a deep-sequencing approach (m<sup>5</sup>C-RIP-seq). LC-MS/MS and dot blot analyses reveal a dynamic pattern of m<sup>5</sup>C mRNA modification in various tissues and at different developmental stages. m<sup>5</sup>C-RIP-seq analysis identified 6045 m<sup>5</sup>C peaks in 4465 expressed genes in young seedlings. We found that m<sup>5</sup>C is enriched in coding sequences with two peaks located immediately after start codons and before stop codons, and is associated with mRNAs with low translation activity. We further demonstrated that an RNA (cytosine-5)-methyltransferase, tRNA-specific methyltransferase 4B (TRM4B), exhibits m<sup>5</sup>C RNA methyltransferase activity. Mutations in TRM4B display defects in root development and decreased m<sup>5</sup>C peaks. TRM4B affects the transcript levels of the genes involved in root development, which is positively correlated with their mRNA stability and m<sup>5</sup>C levels. Our results suggest that m<sup>5</sup>C in mRNA is a new epitranscriptome marker inArabidopsis, and that regulation of this modification is an integral part of gene regulatory networks underlying plant development.</p>',
'date' => '2017-11-06',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28965832',
'doi' => '',
'modified' => '2017-12-04 11:10:34',
'created' => '2017-12-04 11:10:34',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 51 => array(
'id' => '3220',
'name' => 'Maternal obesity programs increased leptin gene expression in rat male offspring via epigenetic modifications in a depot-specific manner',
'authors' => 'Lecoutre S. et al.',
'description' => '<div class="">
<h4>OBJECTIVE:</h4>
<p><abstracttext label="OBJECTIVE" nlmcategory="OBJECTIVE">According to the Developmental Origin of Health and Disease (DOHaD) concept, maternal obesity and accelerated growth in neonates predispose offspring to white adipose tissue (WAT) accumulation. In rodents, adipogenesis mainly develops during lactation. The mechanisms underlying the phenomenon known as developmental programming remain elusive. We previously reported that adult rat offspring from high-fat diet-fed dams (called HF) exhibited hypertrophic adipocyte, hyperleptinemia and increased leptin mRNA levels in a depot-specific manner. We hypothesized that leptin upregulation occurs via epigenetic malprogramming, which takes place early during development of WAT.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">As a first step, we identified <i>in silico</i> two potential enhancers located upstream and downstream of the leptin transcription start site that exhibit strong dynamic epigenomic remodeling during adipocyte differentiation. We then focused on epigenetic modifications (methylation, hydroxymethylation, and histone modifications) of the promoter and the two potential enhancers regulating leptin gene expression in perirenal (pWAT) and inguinal (iWAT) fat pads of HF offspring during lactation (postnatal days 12 (PND12) and 21 (PND21)) and in adulthood.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">PND12 is an active period for epigenomic remodeling in both deposits especially in the upstream enhancer, consistent with leptin gene induction during adipogenesis. Unlike iWAT, some of these epigenetic marks were still observable in pWAT of weaned HF offspring. Retained marks were only visible in pWAT of 9-month-old HF rats that showed a persistent "expandable" phenotype.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Consistent with the DOHaD hypothesis, persistent epigenetic remodeling occurs at regulatory regions especially within intergenic sequences, linked to higher leptin gene expression in adult HF offspring in a depot-specific manner.</abstracttext></p>
</div>',
'date' => '2017-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5518658/',
'doi' => '',
'modified' => '2017-08-18 13:56:40',
'created' => '2017-08-18 13:56:40',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 52 => array(
'id' => '3204',
'name' => 'Increased 5-hydroxymethylation levels in the hippocampus of rat extinguished from cocaine self-administration',
'authors' => 'Sadakierska-Chudy A. et al.',
'description' => '<p>Drug craving and relapse risk during abstinence from cocaine are thought to be caused by persistent changes in transcription and chromatin regulation. Although several brain regions are involved in these processes, the hippocampus seems to play an important role in context-evoked craving and drug-seeking behavior. Only a few studies have examined epigenetic alterations during a period of cocaine abstinence. To investigate the effects of cocaine abstinence on DNA methylation and gene expression, rats that self-administered the drug underwent cocaine abstinence in two time points with extinction training. During the cocaine extinction, we observed elevated global 5-hydroxymethylcytosine(5-hmC) levels with a concurrent increase in Tet3 transcript levels. Moreover, we did not find significant alterations in the levels of Tet3 mRNA and 5-hmC in rats subjected to cocaine abstinence without extinction training. Additionally, our findings demonstrated that the expression of Tet3 target genes was activated. Besides, altered DNA methylation was detected at promoter regions of miRNAs, such as miR-30d and miR-let7i. Further in silico analysis provided evidence that these two molecules targeted the 3' UTR region of the Tet3 gene and thus may contribute to its post-transcriptional regulation. This study has presented novel findings in the hippocampus of rats that underwent extinction training following cocaine self-administration. The alterations in the Tet3 gene expression and the level of 5-hmC may play an important role in extinction learning and the reduction of subsequent cocaine seeking.</p>',
'date' => '2017-07-27',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28422379',
'doi' => '',
'modified' => '2017-07-03 10:21:48',
'created' => '2017-07-03 10:21:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 53 => array(
'id' => '3225',
'name' => 'The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation',
'authors' => 'Leone S. et al.',
'description' => '<p>Long non-coding RNAs (lncRNAs) have been implicated in the regulation of chromatin conformation and epigenetic patterns. lncRNA expression levels are widely taken as an indicator for functional properties. However, the role of RNA processing in modulating distinct features of the same lncRNA is less understood. The establishment of heterochromatin at rRNA genes depends on the processing of IGS-rRNA into pRNA, a reaction that is impaired in embryonic stem cells (ESCs) and activated only upon differentiation. The production of mature pRNA is essential since it guides the repressor TIP5 to rRNA genes, and IGS-rRNA abolishes this process. Through screening for IGS-rRNA-binding proteins, we here identify the RNA helicase DHX9 as a regulator of pRNA processing. DHX9 binds to rRNA genes only upon ESC differentiation and its activity guides TIP5 to rRNA genes and establishes heterochromatin. Remarkably, ESCs depleted of DHX9 are unable to differentiate and this phenotype is reverted by the addition of pRNA, whereas providing IGS-rRNA and pRNA mutants deficient for TIP5 binding are not sufficient. Our results reveal insights into lncRNA biogenesis during development and support a model in which the state of rRNA gene chromatin is part of the regulatory network that controls exit from pluripotency and initiation of differentiation pathways.</p>',
'date' => '2017-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28588071',
'doi' => '',
'modified' => '2017-08-22 13:52:28',
'created' => '2017-08-22 13:52:28',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 54 => array(
'id' => '3233',
'name' => 'Pramel7 mediates ground-state pluripotency through proteasomal-epigenetic combined pathways.',
'authors' => 'Graf U. et al.',
'description' => '<p>Naive pluripotency is established in preimplantation epiblast. Embryonic stem cells (ESCs) represent the immortalization of naive pluripotency. 2i culture has optimized this state, leading to a gene signature and DNA hypomethylation closely comparable to preimplantation epiblast, the developmental ground state. Here we show that Pramel7 (PRAME-like 7), a protein highly expressed in the inner cell mass (ICM) but expressed at low levels in ESCs, targets for proteasomal degradation UHRF1, a key factor for DNA methylation maintenance. Increasing Pramel7 expression in serum-cultured ESCs promotes a preimplantation epiblast-like gene signature, reduces UHRF1 levels and causes global DNA hypomethylation. Pramel7 is required for blastocyst formation and its forced expression locks ESCs in pluripotency. Pramel7/UHRF1 expression is mutually exclusive in ICMs whereas Pramel7-knockout embryos express high levels of UHRF1. Our data reveal an as-yet-unappreciated dynamic nature of DNA methylation through proteasome pathways and offer insights that might help to improve ESC culture to reproduce in vitro the in vivo ground-state pluripotency.</p>',
'date' => '2017-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28604677',
'doi' => '',
'modified' => '2017-08-24 09:57:28',
'created' => '2017-08-24 09:57:28',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 55 => array(
'id' => '3200',
'name' => 'CHD4 Has Oncogenic Functions in Initiating and Maintaining Epigenetic Suppression of Multiple Tumor Suppressor Genes',
'authors' => 'Xia L. et al.',
'description' => '<p>An oncogenic role for CHD4, a NuRD component, is defined for initiating and supporting tumor suppressor gene (TSG) silencing in human colorectal cancer. CHD4 recruits repressive chromatin proteins to sites of DNA damage repair, including DNA methyltransferases where it imposes de novo DNA methylation. At TSGs, CHD4 retention helps maintain DNA hypermethylation-associated transcriptional silencing. CHD4 is recruited by the excision repair protein OGG1 for oxidative damage to interact with the damage-induced base 8-hydroxydeoxyguanosine (8-OHdG), while ZMYND8 recruits it to double-strand breaks. CHD4 knockdown activates silenced TSGs, revealing their role for blunting colorectal cancer cell proliferation, invasion, and metastases. High CHD4 and 8-OHdG levels plus low expression of TSGs strongly correlates with early disease recurrence and decreased overall survival.</p>',
'date' => '2017-05-08',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28486105',
'doi' => '',
'modified' => '2017-07-03 09:56:32',
'created' => '2017-07-03 09:56:32',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 56 => array(
'id' => '3170',
'name' => 'Critical threshold levels of DNA methyltransferase 1 are required to maintain DNA methylation across the genome in human cancer cells.',
'authors' => 'Cai Y. et al.',
'description' => '<p>Reversing DNA methylation abnormalities and associated gene silencing, through inhibiting DNA methyltransferases (DNMTs) is an important potential cancer therapy paradigm. Maximizing this potential requires defining precisely how these enzymes maintain genome-wide, cancer-specific DNA methylation. To date, there is incomplete understanding of precisely how the three DNMTs, 1, 3A, and 3B, interact for maintaining DNA methylation abnormalities in cancer. By combining genetic and shRNA depletion strategies, we define not only a dominant role for DNA methyltransferase 1 (DNMT1) but also distinct roles of 3A and 3B in genome-wide DNA methylation maintenance. Lowering DNMT1 below a threshold level is required for maximal loss of DNA methylation at all genomic regions, including gene body and enhancer regions, and for maximally reversing abnormal promoter DNA hypermethylation and associated gene silencing to reexpress key genes. It is difficult to reach this threshold with patient-tolerable doses of current DNMT inhibitors (DNMTIs). We show that new approaches, like decreasing the DNMT targeting protein, UHRF1, can augment the DNA demethylation capacities of existing DNA methylation inhibitors for fully realizing their therapeutic potential.</p>',
'date' => '2017-04-27',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28232479',
'doi' => '',
'modified' => '2017-05-10 15:31:12',
'created' => '2017-05-10 15:31:12',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 57 => array(
'id' => '3228',
'name' => 'Regulation of DNA demethylation by the XPC DNA repair complex in somatic and pluripotent stem cells.',
'authors' => 'Ho J.J. et al.',
'description' => '<p>Faithful resetting of the epigenetic memory of a somatic cell to a pluripotent state during cellular reprogramming requires DNA methylation to silence somatic gene expression and dynamic DNA demethylation to activate pluripotency gene transcription. The removal of methylated cytosines requires the base excision repair enzyme TDG, but the mechanism by which TDG-dependent DNA demethylation occurs in a rapid and site-specific manner remains unclear. Here we show that the XPC DNA repair complex is a potent accelerator of global and locus-specific DNA demethylation in somatic and pluripotent stem cells. XPC cooperates with TDG genome-wide to stimulate the turnover of essential intermediates by overcoming slow TDG-abasic product dissociation during active DNA demethylation. We further establish that DNA demethylation induced by XPC expression in somatic cells overcomes an early epigenetic barrier in cellular reprogramming and facilitates the generation of more robust induced pluripotent stem cells, characterized by enhanced pluripotency-associated gene expression and self-renewal capacity. Taken together with our previous studies establishing the XPC complex as a transcriptional coactivator, our findings underscore two distinct but complementary mechanisms by which XPC influences gene regulation by coordinating efficient TDG-mediated DNA demethylation along with active transcription during somatic cell reprogramming.</p>',
'date' => '2017-04-05',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28512237',
'doi' => '',
'modified' => '2017-08-23 14:20:13',
'created' => '2017-08-23 14:20:13',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 58 => array(
'id' => '3142',
'name' => 'Epigenetic regulation of RELN and GAD1 in the frontal cortex (FC) of autism spectrum disorder (ASD) subjects',
'authors' => 'Zhubi A. et al.',
'description' => '<p>Both Reelin (RELN) and glutamate decarboxylase 67 (GAD1) have been implicated in the pathophysiology of Autism Spectrum Disorders (ASD). We have previously shown that both mRNAs are reduced in the cerebella (CB) of ASD subjects through a mechanism that involves increases in the amounts of MECP2 binding to the corresponding promoters. In the current study, we examined the expression of RELN, GAD1, GAD2, and several other mRNAs implicated in this disorder in the frontal cortices (FC) of ASD and CON subjects. We also focused on the role that epigenetic processes play in the regulation of these genes in ASD brain. Our goal is to better understand the molecular basis for the down-regulation of genes expressed in GABAergic neurons in ASD brains. We measured mRNA levels corresponding to selected GABAergic genes using qRT-PCR in RNA isolated from both ASD and CON groups. We determined the extent of binding of MECP2 and DNMT1 repressor proteins by chromatin immunoprecipitation (ChIP) assays. The amount of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) present in the promoters of the target genes was quantified by methyl DNA immunoprecipitation (MeDIP) and hydroxyl MeDIP (hMeDIP). We detected significant reductions in the mRNAs associated with RELN and GAD1 and significant increases in mRNAs encoding the Ten-eleven Translocation (TET) enzymes 1, 2, and 3. We also detected increased MECP2 and DNMT1 binding to the corresponding promoter regions of GAD1, RELN, and GAD2. Interestingly, there were decreased amounts of 5mC at both promoters and little change in 5hmC content in these same DNA fragments. Our data demonstrate that RELN, GAD1, and several other genes selectively expressed in GABAergic neurons, are down-regulated in post-mortem ASD FC. In addition, we observed increased DNMT1 and MECP2 binding at the corresponding promoters of these genes. The finding of increased MECP2 binding to the RELN, GAD1 and GAD2 promoters, with reduced amounts of 5mC and unchanged amounts of 5hmC present in these regions, suggests the possibility that DNMT1 interacts with and alters MECP2 binding properties to selected promoters. Comparisons between data obtained from the FC with CB studies showed some common themes between brain regions which are discussed.</p>',
'date' => '2017-02-14',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28229923',
'doi' => '',
'modified' => '2017-03-23 14:58:21',
'created' => '2017-03-23 14:58:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 59 => array(
'id' => '3126',
'name' => 'Intergenerational Transmission of Enhanced Seizure Susceptibility after Febrile Seizures',
'authors' => 'Wu D. et al.',
'description' => '<p>Environmental exposure early in development plays a role in susceptibility to disease in later life. Here, we demonstrate that prolonged febrile seizures induced by exposure of rat pups to a hyperthermic environment enhance seizure susceptibility not only in these hyperthermia-treated rats but also in their future offspring, even if the offspring never experience febrile seizures. This transgenerational transmission was intensity-dependent and was mainly from mothers to their offspring. The transmission was associated with DNA methylation. Thus, our study supports a “Lamarckian”-like mechanism of pathogenesis and the crucial role of epigenetic factors in neurological conditions.</p>',
'date' => '2017-02-08',
'pmid' => 'http://www.sciencedirect.com/science/article/pii/S2352396417300658',
'doi' => '',
'modified' => '2017-02-23 11:05:25',
'created' => '2017-02-23 11:05:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 60 => array(
'id' => '3125',
'name' => 'Pharmacological inhibition of DNA methyltransferase 1 promotes neuronal differentiation from rodent and human nasal olfactory stem/progenitor cell cultures',
'authors' => 'Franco I. et al.',
'description' => '<p>Nasal olfactory stem and neural progenitor cells (NOS/PCs) are considered possible tools for regenerative stem cell therapies in neurodegenerative diseases. Neurogenesis is a complex process regulated by extrinsic and intrinsic signals that include DNA-methylation and other chromatin modifications that could be experimentally manipulated in order to increase neuronal differentiation. The aim of the present study was the characterization of primary cultures and consecutive passages (P2-P10) of NOS/PCs isolated from male Swiss-Webster (mNOS/PCs) or healthy humans (hNOS/PCs). We evaluated and compared cellular morphology, proliferation rates and the expression pattern of pluripotency-associated markers and DNA methylation-associated gene expression in these cultures. Neuronal differentiation was induced by exposure to all-trans retinoic acid and forskolin for 7 days and evaluated by morphological analysis and immunofluorescence against neuronal markers MAP2, NSE and MAP1B. In response to the inductive cues mNOS/PCs expressed NSE (75.67%) and MAP2 (35.34%); whereas the majority of the hNOS/PCs were immunopositive to MAP1B. Treatment with procainamide, a specific inhibitor of DNA methyltransferase 1 (DNMT1), increases in the number of forskolin‘/retinoic acid-induced mature neuronal marker-expressing mNOS/PCs cells and enhances neurite development in hNOS/PCs. Our results indicate that mice and human nasal olfactory stem/progenitors cells share pluripotency-related gene expression suggesting that their application for stem cell therapy is worth pursuing and that DNA methylation inhibitors could be efficient tools to enhance neuronal differentiation from these cells.</p>',
'date' => '2017-02-01',
'pmid' => 'http://www.sciencedirect.com/science/article/pii/S0736574816303665',
'doi' => '',
'modified' => '2017-02-16 10:34:07',
'created' => '2017-02-16 10:34:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 61 => array(
'id' => '3119',
'name' => 'Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain',
'authors' => 'Amort T. et al.',
'description' => '<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec1">
<h3 xmlns="" class="Heading">Background</h3>
<p id="Par1" class="Para">Recent work has identified and mapped a range of posttranscriptional modifications in mRNA, including methylation of the N6 and N1 positions in adenine, pseudouridylation, and methylation of carbon 5 in cytosine (m5C). However, knowledge about the prevalence and transcriptome-wide distribution of m5C is still extremely limited; thus, studies in different cell types, tissues, and organisms are needed to gain insight into possible functions of this modification and implications for other regulatory processes.</p>
</div>
<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec2">
<h3 xmlns="" class="Heading">Results</h3>
<p id="Par2" class="Para">We have carried out an unbiased global analysis of m5C in total and nuclear poly(A) RNA of mouse embryonic stem cells and murine brain. We show that there are intriguing differences in these samples and cell compartments with respect to the degree of methylation, functional classification of methylated transcripts, and position bias within the transcript. Specifically, we observe a pronounced accumulation of m5C sites in the vicinity of the translational start codon, depletion in coding sequences, and mixed patterns of enrichment in the 3′ UTR. Degree and pattern of methylation distinguish transcripts modified in both embryonic stem cells and brain from those methylated in either one of the samples. We also analyze potential correlations between m5C and micro RNA target sites, binding sites of RNA binding proteins, and <em xmlns="" class="EmphasisTypeItalic">N</em>6-methyladenosine.</p>
</div>
<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec3">
<h3 xmlns="" class="Heading">Conclusion</h3>
<p id="Par3" class="Para">Our study presents the first comprehensive picture of cytosine methylation in the epitranscriptome of pluripotent and differentiated stages in the mouse. These data provide an invaluable resource for future studies of function and biological significance of m5C in mRNA in mammals.</p>
</div>',
'date' => '2017-01-05',
'pmid' => 'https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1139-1',
'doi' => '',
'modified' => '2017-02-14 17:20:08',
'created' => '2017-02-14 17:20:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 62 => array(
'id' => '3061',
'name' => 'Novel regional age-associated DNA methylation changes within human common disease-associated loci',
'authors' => 'Bell CG et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">Advancing age progressively impacts on risk and severity of chronic disease. It also modifies the epigenome, with changes in DNA methylation, due to both random drift and variation within specific functional loci.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">In a discovery set of 2238 peripheral-blood genome-wide DNA methylomes aged 19-82 years, we identify 71 age-associated differentially methylated regions within the linkage disequilibrium blocks of the single nucleotide polymorphisms from the NIH genome-wide association study catalogue. This included 52 novel regions, 29 within loci not covered by 450 k or 27 k Illumina array, and with enrichment for DNase-I Hypersensitivity sites across the full range of tissues. These age-associated differentially methylated regions also show marked enrichment for enhancers and poised promoters across multiple cell types. In a replication set of 2084 DNA methylomes, 95.7 % of the age-associated differentially methylated regions showed the same direction of ageing effect, with 80.3 % and 53.5 % replicated to p < 0.05 and p < 1.85 × 10<sup>-8</sup>, respectively.</abstracttext></p>
<h4>CONCLUSION:</h4>
<p><abstracttext label="CONCLUSION" nlmcategory="CONCLUSIONS">By analysing the functionally enriched disease and trait-associated regions of the human genome, we identify novel epigenetic ageing changes, which could be useful biomarkers or provide mechanistic insights into age-related common diseases.</abstracttext></p>
</div>',
'date' => '2016-09-26',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27663977',
'doi' => '',
'modified' => '2016-11-04 10:56:10',
'created' => '2016-11-02 09:54:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 63 => array(
'id' => '3007',
'name' => '5-hydroxymethylcytosine marks postmitotic neural cells in the adult and developing vertebrate central nervous system',
'authors' => 'Diotel N et al.',
'description' => '<p>The epigenetic mark 5-hydroxymethylcytosine (5hmC) is a cytosine modification that is abundant in the central nervous system of mammals and which results from 5-methylcytosine oxidation by TET enzymes. Such a mark is suggested to play key roles in the regulation of chromatin structure and gene expression. However, its precise functions still remain poorly understood and information about its distribution in non-mammalian species is still lacking. Here, the distribution of 5hmC was investigated in the brain of adult zebrafish, African claw frog, and mouse in a comparative manner. We show that zebrafish neurons are endowed with high levels of 5hmC, whereas quiescent or proliferative neural progenitors show low to undetectable levels of the modified cytosine. In the brain of larval and juvenile Xenopus, 5hmC is also detected in neurons, while ventricular proliferative cells do not display this epigenetic mark. Similarly, 5hmC is enriched in neurons compared to neural progenitors of the ventricular zone in the mouse developing cortex. Interestingly, 5hmC colocalized with the methylated DNA binding protein MeCP2 and with the active chromatin histone modification H3K4me2 in mouse neurons. Taken together, our results show an evolutionarily conserved cerebral distribution of 5hmC between fish and tetrapods and reinforce the idea that 5hmC fulfills major functions in the control of chromatin activity in vertebrate neurons.</p>',
'date' => '2016-07-14',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27414756',
'doi' => '',
'modified' => '2016-08-29 09:24:44',
'created' => '2016-08-29 09:24:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 64 => array(
'id' => '2992',
'name' => 'Regulation of the DNA Methylation Landscape in Human Somatic Cell Reprogramming by the miR-29 Family',
'authors' => 'Hysolli E et al.',
'description' => 'Reprogramming to pluripotency after overexpression of OCT4, SOX2, KLF4, and MYC is accompanied by global genomic and epigenomic changes. Histone modification and DNA methylation states in induced pluripotent stem cells (iPSCs) have been shown to be highly similar to embryonic stem cells (ESCs). However, epigenetic differences still exist between iPSCs and ESCs. In particular, aberrant DNA methylation states found in iPSCs are a major concern when using iPSCs in a clinical setting. Thus, it is critical to find factors that regulate DNA methylation states in reprogramming. Here, we found that the miR-29 family is an important epigenetic regulator during human somatic cell reprogramming. Our global DNA methylation and hydroxymethylation analysis shows that DNA demethylation is a major event mediated by miR-29a depletion during early reprogramming, and that iPSCs derived from miR-29a depletion are epigenetically closer to ESCs. Our findings uncover an important miRNA-based approach to generate clinically robust iPSCs.',
'date' => '2016-07-12',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27373925',
'doi' => '10.1016/j.stemcr.2016.05.014',
'modified' => '2016-08-23 09:57:29',
'created' => '2016-08-23 09:57:29',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 65 => array(
'id' => '3011',
'name' => 'Genome-Wide DNA Methylation in Mixed Ancestry Individuals with Diabetes and Prediabetes from South Africa',
'authors' => 'Matsha TE et al.',
'description' => '<p>Aims. To conduct a genome-wide DNA methylation in individuals with type 2 diabetes, individuals with prediabetes, and control mixed ancestry individuals from South Africa. Methods. We used peripheral blood to perform genome-wide DNA methylation analysis in 3 individuals with screen detected diabetes, 3 individuals with prediabetes, and 3 individuals with normoglycaemia from the Bellville South Community, Cape Town, South Africa, who were age-, gender-, body mass index-, and duration of residency-matched. Methylated DNA immunoprecipitation (MeDIP) was performed by Arraystar Inc. (Rockville, MD, USA). Results. Hypermethylated DMRs were 1160 (81.97%) and 124 (43.20%), respectively, in individuals with diabetes and prediabetes when both were compared to subjects with normoglycaemia. Our data shows that genes related to the immune system, signal transduction, glucose transport, and pancreas development have altered DNA methylation in subjects with prediabetes and diabetes. Pathway analysis based on the functional analysis mapping of genes to KEGG pathways suggested that the linoleic acid metabolism and arachidonic acid metabolism pathways are hypomethylated in prediabetes and diabetes. Conclusions. Our study suggests that epigenetic changes are likely to be an early process that occurs before the onset of overt diabetes. Detailed analysis of DMRs that shows gradual methylation differences from control versus prediabetes to prediabetes versus diabetes in a larger sample size is required to confirm these findings.</p>',
'date' => '2016-06-28',
'pmid' => 'http://www.hindawi.com/journals/ije/2016/3172093/',
'doi' => '',
'modified' => '2016-08-29 10:27:14',
'created' => '2016-08-29 10:27:14',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 66 => array(
'id' => '2954',
'name' => 'Dnmt2/Trdmt1 as Mediator of RNA Polymerase II Transcriptional Activity in Cardiac Growth',
'authors' => 'Ghanbarian H et al.',
'description' => '<p>Dnmt2/Trdmt1 is a methyltransferase, which has been shown to methylate tRNAs. Deficient mutants were reported to exhibit various, seemingly unrelated, defects in development and RNA-mediated epigenetic heredity. Here we report a role in a distinct developmental regulation effected by a noncoding RNA. We show that Dnmt2-deficiency in mice results in cardiac hypertrophy. Echocardiographic measurements revealed that cardiac function is preserved notwithstanding the increased dimensions of the organ due to cardiomyocyte enlargement. Mechanistically, activation of the P-TEFb complex, a critical step for cardiac growth, results from increased dissociation of the negatively regulating Rn7sk non-coding RNA component in Dnmt2-deficient cells. Our data suggest that Dnmt2 plays an unexpected role for regulation of cardiac growth by modulating activity of the P-TEFb complex.</p>',
'date' => '2016-06-06',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27270731',
'doi' => ' 10.1371/journal.pone.0156953',
'modified' => '2016-06-14 15:49:17',
'created' => '2016-06-14 15:49:17',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 67 => array(
'id' => '2941',
'name' => 'Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers',
'authors' => 'Li L et al.',
'description' => '<p>Promoter CpG methylation is a fundamental regulatory process of gene expression. TET proteins are active CpG demethylases converting 5-methylcytosine to 5-hydroxymethylcytosine, with loss of 5 hmC as an epigenetic hallmark of cancers, indicating critical roles of TET proteins in epigenetic tumorigenesis. Through analysis of tumor methylomes, we discovered <i>TET1</i> as a methylated target, and further confirmed its frequent downregulation/methylation in cell lines and primary tumors of multiple carcinomas and lymphomas, including nasopharyngeal, esophageal, gastric, colorectal, renal, breast and cervical carcinomas, as well as non-Hodgkin, Hodgkin and nasal natural killer/T-cell lymphomas, although all three <i>TET</i> family genes are ubiquitously expressed in normal tissues. Ectopic expression of TET1 catalytic domain suppressed colony formation and induced apoptosis of tumor cells of multiple tissue types, supporting its role as a broad <i>bona fide</i> tumor suppressor. Furthermore, TET1 catalytic domain possessed demethylase activity in cancer cells, being able to inhibit the CpG methylation of tumor suppressor gene (TSG) promoters and reactivate their expression, such as <i>SLIT2, ZNF382</i> and <i>HOXA9</i>. As only infrequent mutations of <i>TET1</i> have been reported, compared to <i>TET2</i>, epigenetic silencing therefore appears to be the dominant mechanism for <i>TET1</i> inactivation in cancers, which also forms a feedback loop of CpG methylation during tumorigenesis.</p>',
'date' => '2016-05-26',
'pmid' => 'http://www.nature.com/articles/srep26591',
'doi' => '10.1038/srep26591',
'modified' => '2016-06-06 09:47:31',
'created' => '2016-06-06 09:47:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 68 => array(
'id' => '2836',
'name' => 'Biochemical reconstitution of TET1–TDG–BER-dependent active DNA demethylation reveals a highly coordinated mechanism',
'authors' => 'Weber AR, Krawczyk C, Robertson AB, Kuśnierczyk A, Vågbø CB, Schuermann D, Klungland A, Schär P',
'description' => '<p>Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten–eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET–TDG–BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.nature.com/ncomms/2016/160302/ncomms10806/full/ncomms10806.html',
'doi' => '10.1038/ncomms10806',
'modified' => '2016-03-08 10:25:46',
'created' => '2016-03-08 10:25:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 69 => array(
'id' => '2837',
'name' => 'Hydroxymethylation of microRNA-365-3p Regulates Nociceptive Behaviors via Kcnh2',
'authors' => 'Pan Z, Zhang M, Ma T, Xue Z-Y, Li G-F, Hao L-Y, Zhu L-J, Li Y-Q, Ding H-L, Cao J-L',
'description' => '<p id="p-3">DNA 5-hydroxylmethylcytosine (5hmC) catalyzed by ten-eleven translocation methylcytosine dioxygenase (TET) occurs abundantly in neurons of mammals. However, the <em>in vivo</em> causal link between TET dysregulation and nociceptive modulation has not been established. Here, we found that spinal TET1 and TET3 were significantly increased in the model of formalin-induced acute inflammatory pain, which was accompanied with the augment of genome-wide 5hmC content in spinal cord. Knockdown of spinal TET1 or TET3 alleviated the formalin-induced nociceptive behavior and overexpression of spinal TET1 or TET3 in naive mice produced pain-like behavior as evidenced by decreased thermal pain threshold. Furthermore, we found that TET1 or TET3 regulated the nociceptive behavior by targeting microRNA-365-3p (miR-365-3p). Formalin increased 5hmC in the miR-365-3p promoter, which was inhibited by knockdown of TET1 or TET3 and mimicked by overexpression of TET1 or TET3 in naive mice. Nociceptive behavior induced by formalin or overexpression of spinal TET1 or TET3 could be prevented by downregulation of miR-365-3p, and mimicked by overexpression of spinal miR-365-3p. Finally, we demonstrated that a potassium channel, voltage-gated eag-related subfamily H member 2 (<em>Kcnh2</em>), validated as a target of miR-365-3p, played a critical role in nociceptive modulation by spinal TET or miR-365-3p. Together, we concluded that TET-mediated hydroxymethylation of miR-365-3p regulates nociceptive behavior via <em>Kcnh2</em>.</p>
<p id="p-4"><strong>SIGNIFICANCE STATEMENT</strong> Mounting evidence indicates that epigenetic modifications in the nociceptive pathway contribute to pain processes and analgesia response. Here, we found that the increase of 5hmC content mediated by TET1 or TET3 in miR-365-3p promoter in the spinal cord is involved in nociceptive modulation through targeting a potassium channel, <em>Kcnh2</em>. Our study reveals a new epigenetic mechanism underlying nociceptive information processing, which may be a novel target for development of antinociceptive drugs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.jneurosci.org/content/36/9/2769.short',
'doi' => '10.1523/JNEUROSCI.3474-15.2016',
'modified' => '2016-03-08 10:40:48',
'created' => '2016-03-08 10:40:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 70 => array(
'id' => '3034',
'name' => 'Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism',
'authors' => 'Weber AR et al.',
'description' => '<p>Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten-eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET-TDG-BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26932196',
'doi' => '',
'modified' => '2016-09-23 16:34:57',
'created' => '2016-09-23 16:34:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 71 => array(
'id' => '2833',
'name' => 'Genome-wide DNA methylation profile of developing deciduous tooth germ in miniature pigs',
'authors' => 'Su Y, Fan Z, Wu X, Li Y, Wang F, Zhang C, Wang J, Du J, Wang S',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND"><span class="highlight">DNA</span> <span class="highlight">methylation</span> is an important epigenetic modification critical to the regulation of gene expression during development. To date, little is known about the role of <span class="highlight">DNA</span> <span class="highlight">methylation</span> in <span class="highlight">tooth</span> development in large animal models. Thus, we carried out a comparative genomic analysis of <span class="highlight">genome-wide</span> <span class="highlight">DNA</span> <span class="highlight">methylation</span> profiles in E50 and E60 <span class="highlight">tooth</span> <span class="highlight">germ</span> from <span class="highlight">miniature</span> <span class="highlight">pigs</span> using methylated <span class="highlight">DNA</span> immunoprecipitation-sequencing (MeDIP-seq).</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We observed different <span class="highlight">DNA</span> <span class="highlight">methylation</span> patterns during the different developmental stages of pig <span class="highlight">tooth</span> <span class="highlight">germ</span>. A total of 2469 differentially methylated genes were identified. Functional analysis identified several signaling pathways and 104 genes that may be potential key regulators of pig <span class="highlight">tooth</span> development from E50 to E60.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">The present study provided a comprehensive analysis of the global <span class="highlight">DNA</span> <span class="highlight">methylation</span> pattern of <span class="highlight">tooth</span> <span class="highlight">germ</span> in <span class="highlight">miniature</span> <span class="highlight">pigs</span> and identified candidate genes that potentially regulate <span class="highlight">tooth</span> development from E50 to E60.</abstracttext></p>
</div>',
'date' => '2016-02-24',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26911717',
'doi' => '10.1186/s12864-016-2485-9',
'modified' => '2016-03-01 10:35:04',
'created' => '2016-03-01 10:35:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 72 => array(
'id' => '2834',
'name' => 'Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.',
'authors' => 'Rebollo R, Mager DL',
'description' => '<p>Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method used in our laboratory to detect, quantify, and compare mammalian endogenous retrovirus DNA methylation. More specifically we describe methylated DNA immunoprecipitation (MeDIP) followed by quantitative PCR.</p>',
'date' => '2016-02-20',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26895065',
'doi' => '10.1007/978-1-4939-3372-3_23',
'modified' => '2016-03-02 10:37:15',
'created' => '2016-03-02 10:22:16',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 73 => array(
'id' => '2815',
'name' => 'The dual specificity phosphatase 2 gene is hypermethylated in human cancer and regulated by epigenetic mechanisms',
'authors' => 'Tanja Haag, Antje M. Richter, Martin B. Schneider, Adriana P. Jiménez and Reinhard H. Dammann',
'description' => '<p><span>Dual specificity phosphatases are a class of tumor-associated proteins involved in the negative regulation of the MAP kinase pathway. Downregulation of the </span><em xmlns="" class="EmphasisTypeItalic">dual specificity phosphatase 2</em><span> (</span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span>) has been reported in cancer. Epigenetic silencing of tumor suppressor genes by abnormal promoter methylation is a frequent mechanism in oncogenesis. It has been shown that the epigenetic factor CTCF is involved in the regulation of tumor suppressor genes.</span></p>
<p><span>We analyzed the promoter hypermethylation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> in human cancer, including primary Merkel cell carcinoma by bisulfite restriction analysis and pyrosequencing. Moreover we analyzed the impact of a DNA methyltransferase inhibitor (5-Aza-dC) and CTCF on the epigenetic regulation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> by qRT-PCR, promoter assay, chromatin immuno-precipitation and methylation analysis.</span></span></p>
<p><span><span>Here we report a significant tumor-specific hypermethylation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> in primary Merkel cell carcinoma (</span><em xmlns="" class="EmphasisTypeItalic">p</em><span> = 0.05). An increase in methylation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> was also found in 17 out of 24 (71 %) cancer cell lines, including skin and lung cancer. Treatment of cancer cells with 5-Aza-dC induced </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression by its promoter demethylation, Additionally we observed that CTCF induces </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression in cell lines that exhibit silencing of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span>. This reactivation was accompanied by increased CTCF binding and demethylation of the </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> promoter.</span></span></span></p>
<p><span><span><span>Our data show that aberrant epigenetic inactivation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> occurs in carcinogenesis and that CTCF is involved in the epigenetic regulation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression.</span></span></span></span></p>',
'date' => '2016-02-01',
'pmid' => 'http://pubmed.gov/26833217',
'doi' => '10.1186/s12885-016-2087-6',
'modified' => '2016-03-09 16:34:51',
'created' => '2016-02-09 14:47:36',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 74 => array(
'id' => '2859',
'name' => 'Role of Growth Arrest and DNA Damage-Inducible, Beta in Alcohol-Drinking Behaviors',
'authors' => 'Gavin DP, Kusumo H, Zhang H, Guidotti A, Pandey SC',
'description' => '<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">The contribution of epigenetic factors, such as histone acetylation and DNA methylation, to the regulation of alcohol-drinking behavior has been increasingly recognized over the last several years. GADD45b is a protein demonstrated to be involved in DNA demethylation at neurotrophic factor gene promoters, including at brain-derived neurotrophic factor (Bdnf) which has been highly implicated in alcohol-drinking behavior.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">DNA methyltransferase-1 (Dnmt1), 3a, and 3b, and Gadd45a, b, and g mRNA were measured in the nucleus accumbens (NAc) and ventral tegmental areas of high ethanol (EtOH) consuming C57BL/6J (C57) and low alcohol consuming DBA/2J (DBA) mice using quantitative reverse transcriptase polymerase chain reaction (PCR). In the NAc, GADD45b protein was measured via immunohistochemistry and Bdnf9a mRNA using in situ PCR. Bdnf9a promoter histone H3 acetylated at lysines 9 and 14 (H3K9,K14ac) was measured using chromatin immunoprecipitation, and 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) using methylated DNA immunoprecipitation. Alcohol-drinking behavior was evaluated in Gadd45b haplodeficient (+/-) and null mice (-/-) utilizing drinking-in-the-dark (DID) and 2-bottle free-choice paradigms.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">C57 mice had lower levels of Gadd45b and g mRNA and GADD45b protein in the NAc relative to the DBA strain. C57 mice had lower NAc shell Bdnf9a mRNA levels, Bdnf9a promoter H3K9,K14ac, and higher Bdnf9a promoter 5HMC and 5MC. Acute EtOH increased GADD45b protein, Bdnf9a mRNA, and histone acetylation and decreased 5HMC in C57 mice. Gadd45b +/- mice displayed higher drinking behavior relative to wild-type littermates in both DID and 2-bottle free-choice paradigms.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">These data indicate the importance of the DNA demethylation pathway and its interactions with histone posttranslational modifications in alcohol-drinking behavior. Further, we suggest that lower DNA demethylation protein GADD45b levels may affect Bdnf expression possibly leading to altered alcohol-drinking behavior.</abstracttext></p>',
'date' => '2016-02-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26842245',
'doi' => ' 10.1111/acer.12965',
'modified' => '2016-03-15 16:37:22',
'created' => '2016-03-15 16:37:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 75 => array(
'id' => '2975',
'name' => 'Protocol for Methylated DNA Immunoprecipitation (MeDIP) Analysis',
'authors' => 'Karpova NN et al.',
'description' => '<p>DNA methylation is a fundamental epigenetic mechanism for silencing gene expression by either modifying chromatin structure to a repressive state or interfering with the transcription factors’ binding. DNA methylation primarily occurs at the position C5 of a cytosine ring mainly in the context of CpG dinucleotides. The modification can be recognized both in vivo and in vitro by the methyl-CpG binding proteins (MBPs) as well as in vitro by an antibody raised against 5-methylcytosine (5mC). This chapter describes different MBPs and introduces a standard methylated DNA immunoprecipitation (MeDIP) method, which is based on using the anti-5mC antibody to isolate methylated DNA fragments for subsequent locus-specific DNA methylation analysis. The MeDIP-generated DNA can be used as well for methylation profiling on a genome scale using array-based (MeDIP-chip) and high-throughput (MeDIP-seq) technologies.</p>',
'date' => '2016-02-01',
'pmid' => 'http://link.springer.com/protocol/10.1007/978-1-4939-2754-8_6',
'doi' => '10.1007/978-1-4939-2754-8_6',
'modified' => '2016-07-07 09:35:44',
'created' => '2016-07-07 09:35:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 76 => array(
'id' => '2844',
'name' => 'De novo DNA methylation drives 5hmC accumulation in mouse zygotes',
'authors' => 'Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, Koseki H, Sasaki H, Hajkova P',
'description' => '<p>Zygotic epigenetic reprogramming entails genome-wide DNA demethylation that is accompanied by Tet methylcytosine dioxygenase 3 (Tet3)-driven oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC; refs ,,,). Here we demonstrate using detailed immunofluorescence analysis and ultrasensitive LC-MS-based quantitative measurements that the initial loss of paternal 5mC does not require 5hmC formation. Small-molecule inhibition of Tet3 activity, as well as genetic ablation, impedes 5hmC accumulation in zygotes without affecting the early loss of paternal 5mC. Instead, 5hmC accumulation is dependent on the activity of zygotic Dnmt3a and Dnmt1, documenting a role for Tet3-driven hydroxylation in targeting de novo methylation activities present in the early embryo. Our data thus provide further insights into the dynamics of zygotic reprogramming, revealing an intricate interplay between DNA demethylation, de novo methylation and Tet3-driven hydroxylation.</p>',
'date' => '2016-01-11',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26751286',
'doi' => '10.1038/ncb3296',
'modified' => '2016-03-09 17:32:33',
'created' => '2016-03-09 17:32:33',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 77 => array(
'id' => '2860',
'name' => 'DNA methylation profiling: comparison of genome-wide sequencing methods and the Infinium Human Methylation 450 Bead Chip',
'authors' => 'Walker DL, Bhagwate AV, Baheti S, Smalley RL, Hilker CA, Sun Z, Cunningham JM',
'description' => '<div class="">
<h4>AIMS:</h4>
<p><abstracttext label="AIMS" nlmcategory="OBJECTIVE">To compare the performance of four sequence-based and one microarray methods for DNA methylation profiling.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">DNA from two cell lines were profiled by reduced representation bisulfite sequencing, methyl capture sequencing (SS-Meth Seq), NimbleGen SeqCapEpi CpGiant(Nimblegen MethSeq), methylated DNA immunoprecipitation (MeDIP) and the Human Methylation 450 Bead Chip (Meth450K).</abstracttext></p>
<h4>RESULTS & CONCLUSION:</h4>
<p><abstracttext label="RESULTS & CONCLUSION" nlmcategory="CONCLUSIONS">Despite differences in genome-wide coverage, high correlation and concordance were observed between different methods. Significant overlap of differentially methylated regions was identified between sequenced-based platforms. MeDIP provided the best coverage for the whole genome and gene body regions, while RRBS and Nimblegen MethSeq were superior for CpGs in CpG islands and promoters. Methylation analyses can be achieved by any of the five methods but understanding their differences may better address the research question being posed.</abstracttext></p>
</div>',
'date' => '2015-12-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26192535',
'doi' => '10.2217/EPI.15.64',
'modified' => '2016-03-16 11:06:05',
'created' => '2016-03-16 11:06:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 78 => array(
'id' => '2931',
'name' => 'Oxidative DNA damage in mouse sperm chromosomes: Size matters.',
'authors' => 'Kocer A et al.',
'description' => '<p>Normal embryo and foetal development as well as the health of the progeny are mostly dependent on gamete nuclear integrity. In the present study, in order to characterize more precisely oxidative DNA damage in mouse sperm we used two mouse models that display high levels of sperm oxidative DNA damage, a common alteration encountered both in in vivo and in vitro reproduction. Immunoprecipitation of oxidized sperm DNA coupled to deep sequencing showed that mouse chromosomes may be largely affected by oxidative alterations. We show that the vulnerability of chromosomes to oxidative attack inversely correlated with their size and was not linked to their GC richness. It was neither correlated with the chromosome content in persisting nucleosomes nor associated with methylated sequences. A strong correlation was found between oxidized sequences and sequences rich in short interspersed repeat elements (SINEs). Chromosome position in the sperm nucleus as revealed by fluorescent in situ hybridization appears to be a confounder. These data map for the first time fragile mouse sperm chromosomal regions when facing oxidative damage that may challenge the repair mechanisms of the oocyte post-fertilization.</p>',
'date' => '2015-12-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26510519',
'doi' => '10.1016/j.freeradbiomed.2015.10.419',
'modified' => '2016-05-19 10:18:24',
'created' => '2016-05-19 10:18:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 79 => array(
'id' => '2944',
'name' => 'Immunohistochemical Detection of Oxidized Forms of 5-Methylcytosine in Embryonic and Adult Brain Tissue',
'authors' => 'Abakir A et al.',
'description' => '<p>DNA methylation (5-methylcytosine, 5mC) is a major epigenetic modification of the eukaryotic genome associated with gene repression. Ten-eleven translocation proteins (Tet1/2/3) can oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Recent studies demonstrate that 5hmC is particularly enriched in neuronal cells and imply the involvement of this mark in transcriptional regulation taking place within the mammalian brain. Although a number of biochemical and antibody-based approaches have been successfully used to study the global content and genomic distributions of 5hmC in various contexts, most of these techniques do not provide any spatial information on the levels of this mark in different cell types. Here we describe a method of sensitive immunochemical detection of 5hmC/5fC/5caC in brain tissue based on the use of peroxidase-conjugated secondary antibodies and tyramide signal amplification. This technique can be instrumental in determining the relative enrichments of oxidized forms of 5mC in different brain cell types, effectively complementing other established approaches to investigate the functions of these marks in embryonic and adult brain.</p>',
'date' => '2015-09-02',
'pmid' => 'http://link.springer.com/protocol/10.1007%2F978-1-4939-2754-8_8',
'doi' => ' Print ISBN 978-1-4939-2753-1 Online ISBN 978-1-4939-2754-8 Series Title Neuromethods Series Volume 105 Series ISSN 0893-2336 Publisher Springer New York Copyright Holder Springer Science+Business Media New York Additional Lin',
'modified' => '2016-06-08 10:16:17',
'created' => '2016-06-08 10:16:17',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 80 => array(
'id' => '2847',
'name' => 'Gadd45b and N-methyl-D-aspartate induced DNA demethylation in postmitotic neurons.',
'authors' => 'Gavin DP, Kusumo H, Sharma RP, Guizzetti M, Guidotti A, Pandey SC.',
'description' => '<p><strong>AIM:</strong> In nondividing neurons examine the role of Gadd45b in active 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) removal at a gene promoter highly implicated in mental illnesses and cognition, Bdnf.</p>
<p><strong>MATERIALS & METHODS:</strong> Mouse primary cortical neuronal cultures with and without Gadd45b siRNA transfection were treated with N-methyl-d-aspartate (NMDA). Expression changes of genes reportedly involved in DNA demethylation, Bdnf mRNA and protein and 5MC and 5HMC at Bdnf promoters were measured.</p>
<p><strong>RESULTS:</strong> Gadd45b siRNA transfection in neurons abolishes the NMDA-induced increase in Bdnf IXa mRNA and reductions in 5MC and 5HMC at the Bdnf IXa promoter.</p>
<p><strong>CONCLUSION:</strong> These results contribute to our understanding of DNA demethylation mechanisms in neurons, and its role in regulating NMDA responsive genes implicated in mental illnesses.</p>',
'date' => '2015-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/26111030',
'doi' => '10.2217/epi.15.12',
'modified' => '2016-03-11 16:02:08',
'created' => '2016-03-11 15:47:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 81 => array(
'id' => '2879',
'name' => 'Active human nucleolar organizer regions are interspersed with inactive rDNA repeats in normal and tumor cells.',
'authors' => 'Zillner K, Komatsu J, Filarsky K, Kalepu R, Bensimon A, Németh A',
'description' => '<div class="">
<h4>AIM:</h4>
<p><abstracttext label="AIM" nlmcategory="OBJECTIVE">The synthesis of rRNA is a key determinant of normal and malignant cell growth and subject to epigenetic regulation. Yet, the epigenomic features of rDNA arrays clustered in nucleolar organizer regions are largely unknown. We set out to explore for the first time how DNA methylation is distributed on individual rDNA arrays.</abstracttext></p>
<h4>MATERIALS & METHODS:</h4>
<p><abstracttext label="MATERIALS & METHODS" nlmcategory="METHODS">Here we combined immunofluorescence detection of DNA modifications with fluorescence hybridization of single DNA fibers, metaphase immuno-FISH and methylation-sensitive restriction enzyme digestions followed by Southern blot.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We found clustering of both hypomethylated and hypermethylated repeat units and hypermethylation of noncanonical rDNA in IMR90 fibroblasts and HCT116 colorectal carcinoma cells. Surprisingly, we also found transitions between hypo- and hypermethylated rDNA repeat clusters on single DNA fibers.</abstracttext></p>
<h4>CONCLUSION:</h4>
<p><abstracttext label="CONCLUSION" nlmcategory="CONCLUSIONS">Collectively, our analyses revealed co-existence of different epialleles on individual nucleolar organizer regions and showed that epi-combing is a valuable approach to analyze epigenomic patterns of repetitive DNA.</abstracttext></p>
</div>',
'date' => '2015-06-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26077426',
'doi' => '10.2217/epi.14.93',
'modified' => '2016-04-05 09:44:29',
'created' => '2016-04-05 09:44:29',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 82 => array(
'id' => '2790',
'name' => 'Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency.',
'authors' => 'Chen H, Aksoy I, Gonnot F, Osteil P, Aubry M, Hamela C, Rognard C, Hochard A, Voisin S, Fontaine E, Mure M, Afanassieff M, Cleroux E, Guibert S, Chen J, Vallot C, Acloque H, Genthon C, Donnadieu C, De Vos J, Sanlaville D, Guérin JF, Weber M, Stanton LW, R',
'description' => 'Leukemia inhibitory factor (LIF)/STAT3 signalling is a hallmark of naive pluripotency in rodent pluripotent stem cells (PSCs), whereas fibroblast growth factor (FGF)-2 and activin/nodal signalling is required to sustain self-renewal of human PSCs in a condition referred to as the primed state. It is unknown why LIF/STAT3 signalling alone fails to sustain pluripotency in human PSCs. Here we show that the forced expression of the hormone-dependent STAT3-ER (ER, ligand-binding domain of the human oestrogen receptor) in combination with 2i/LIF and tamoxifen allows human PSCs to escape from the primed state and enter a state characterized by the activation of STAT3 target genes and long-term self-renewal in FGF2- and feeder-free conditions. These cells acquire growth properties, a gene expression profile and an epigenetic landscape closer to those described in mouse naive PSCs. Together, these results show that temporarily increasing STAT3 activity is sufficient to reprogramme human PSCs to naive-like pluripotent cells.',
'date' => '2015-05-13',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25968054',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 83 => array(
'id' => '2678',
'name' => 'Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells.',
'authors' => 'Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, Pop R, Reyon D, Tsai SQ, Mallard W, Joung JK, Rinn JL, Gnirke A, Meissner A',
'description' => 'DNA methylation is a key epigenetic modification involved in regulating gene expression and maintaining genomic integrity. Here we inactivated all three catalytically active DNA methyltransferases (DNMTs) in human embryonic stem cells (ESCs) using CRISPR/Cas9 genome editing to further investigate the roles and genomic targets of these enzymes. Disruption of DNMT3A or DNMT3B individually as well as of both enzymes in tandem results in viable, pluripotent cell lines with distinct effects on the DNA methylation landscape, as assessed by whole-genome bisulfite sequencing. Surprisingly, in contrast to findings in mouse, deletion of DNMT1 resulted in rapid cell death in human ESCs. To overcome this immediate lethality, we generated a doxycycline-responsive tTA-DNMT1* rescue line and readily obtained homozygous DNMT1-mutant lines. However, doxycycline-mediated repression of exogenous DNMT1* initiates rapid, global loss of DNA methylation, followed by extensive cell death. Our data provide a comprehensive characterization of DNMT-mutant ESCs, including single-base genome-wide maps of the targets of these enzymes.',
'date' => '2015-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25822089',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 84 => array(
'id' => '2477',
'name' => 'Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes.',
'authors' => 'Li L, Zhang Y, Fan Y, Sun K, Su X, Du Z, Tsao SW, Loh TK, Sun H, Chan AT, Zeng YX, Chan WY, Chan FK, Tao Q',
'description' => 'Aims: Nasopharyngeal carcinoma (NPC) is a common tumor consistently associated with Epstein-Barr virus infection and prevalent in South China, including Hong Kong, and southeast Asia. Current genomic sequencing studies found only rare mutations in NPC, indicating its critical epigenetic etiology, while no epigenome exists for NPC as yet. Materials & methods: We profiled the methylomes of NPC cell lines and primary tumors, together with normal nasopharyngeal epithelial cells, using methylated DNA immunoprecipitation (MeDIP). Results: We observed extensive, genome-wide methylation of cellular genes. Epigenetic disruption of Wnt, MAPK, TGF-β and Hedgehog signaling pathways was detected. Methylation of Wnt signaling regulators (SFRP1, 2, 4 and 5, DACT2, DKK2 and DKK3) was frequently detected in tumor and nasal swab samples from NPC patients. Functional studies showed that these genes are bona fide tumor-suppressor genes for NPC. Conclusion: The NPC methylome shows a special high-degree CpG methylation epigenotype, similar to the Epstein-Barr virus-infected gastric cancer, indicating a critical epigenetic etiology for NPC pathogenesis.',
'date' => '2015-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25479246',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 85 => array(
'id' => '2613',
'name' => 'CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.',
'authors' => 'Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC',
'description' => 'BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the underlying epigenetic signatures involved in its regulation. RESULTS: NOS3 and STAT3 mRNA levels were elevated in HUAEC of patients who suffered from placental insufficiency. 5-hydroxymethylcytosine, H3K9ac and Histone 2A (H2A).Zac at the NOS3 transcription start site directly correlated with NOS3 mRNA levels. Concomitantly, we observed entangled histone acetylation patterns and NOS3 response upon hypoxic conditions in vitro. Knock-down of either NOS3 or STAT3 by RNAi provided evidence for a functional NOS3/STAT3 relationship. Moreover, we recognized massive turnover of Stat3 at a discrete binding site in the NOS3 promoter. Interestingly, induced hyperacetylation resulted in short-termed increase of NOS3 mRNA followed by deferred decrease indicating that NOS3 expression could become self-attenuated by co-expressed intronic 27 nt-ncRNA. Reporter assay results and phylogenetic analyses enabled us to propose a novel model for STAT3-3'-UTR targeting by this 27-nt-ncRNA. CONCLUSIONS: An adverse intrauterine environment leads to adaptive changes of NOS3 expression. Apparently, a rapid NOS3 self-limiting response upon ectopic triggers co-exists with longer termed expression changes in response to placental insufficiency involving differential epigenetic signatures. Their persistence might contribute to impaired vascular endothelial response and consequently increase the risk of cardiovascular disease later in life.',
'date' => '2015-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25699114',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 86 => array(
'id' => '2437',
'name' => 'Acute Depletion Redefines the Division of Labor among DNA Methyltransferases in Methylating the Human Genome.',
'authors' => 'Tiedemann RL, Putiri EL, Lee JH, Hlady RA, Kashiwagi K, Ordog T, Zhang Z, Liu C, Choi JH, Robertson KD',
'description' => 'Global patterns of DNA methylation, mediated by the DNA methyltransferases (DNMTs), are disrupted in all cancers by mechanisms that remain largely unknown, hampering their development as therapeutic targets. Combinatorial acute depletion of all DNMTs in a pluripotent human tumor cell line, followed by epigenome and transcriptome analysis, revealed DNMT functions in fine detail. DNMT3B occupancy regulates methylation during differentiation, whereas an unexpected interplay was discovered in which DNMT1 and DNMT3B antithetically regulate methylation and hydroxymethylation in gene bodies, a finding confirmed in other cell types. DNMT3B mediated non-CpG methylation, whereas DNMT3L influenced the activity of DNMT3B toward non-CpG versus CpG site methylation. Altogether, these data reveal functional targets of each DNMT, suggesting that isoform selective inhibition would be therapeutically advantageous.',
'date' => '2014-11-20',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25453758',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 87 => array(
'id' => '2368',
'name' => 'A B-cell targeting virus disrupts potentially protective genomic methylation patterns in lymphoid tissue by increasing global 5-hydroxmethylcytosine levels',
'authors' => 'Ciccone NA, Mwangi W, Ruzov A, Smith LP, Butter C, Nair V',
'description' => 'The mechanisms by which viruses modulate the immune system include changes in host genomic methylation. 5-hydroxmethylcytosine (5hmC) is the catalytic product of the Tet (Ten-11 translocation) family of enzymes and may serve as an intermediate of DNA demethylation. Recent reports suggest that 5hmC may confer consequences on cellular events including the pathogenesis of disease; in order to explore this possibility further we investigated both 5-methylcytosine (5mC) and 5hmC levels in healthy and diseased chicken bursas of Fabricius. We discovered that embryonic B-cells have high 5mC content while 5hmC decreases during bursa development. We propose that a high 5mC level protects from the mutagenic activity of the B-cell antibody diversifying enzyme activation induced deaminase (AID). In support of this view, AID mRNA increases significantly within the developing bursa from embryonic to post hatch stages while mRNAs that encode Tet family members 1 and 2 reduce over the same period. Moreover, our data revealed that infectious bursal disease virus (IBDV) disrupts this genomic methylation pattern causing a global increase in 5hmC levels in a mechanism that may involve increased Tet 1 and 2 mRNAs. To our knowledge this is the first time that a viral infection has been observed to cause global increases in genomic 5hmC within infected host tissues, underlining a mechanism that may involve the induction of B-cell genomic instability and cell death to facilitate viral egress.',
'date' => '2014-10-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/25338704',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 88 => array(
'id' => '2350',
'name' => 'Spontaneous sleep-wake cycle and sleep deprivation differently induce Bdnf1, Bdnf4 and Bdnf9a DNA methylation and transcripts levels in the basal forebrain and frontal cortex in rats.',
'authors' => 'Ventskovska O, Porkka-Heiskanen T, Karpova NN',
'description' => 'Brain-derived neurotrophic factor (Bdnf) regulates neuronal plasticity, slow wave activity and sleep homeostasis. Environmental stimuli control Bdnf expression through epigenetic mechanisms, but there are no data on epigenetic regulation of Bdnf by sleep or sleep deprivation. Here we investigated whether 5-methylcytosine (5mC) DNA modification at Bdnf promoters p1, p4 and p9 influences Bdnf1, Bdnf4 and Bdnf9a expression during the normal inactive phase or after sleep deprivation (SD) (3, 6 and 12 h, end-times being ZT3, ZT6 and ZT12) in rats in two brain areas involved in sleep regulation, the basal forebrain and cortex. We found a daytime variation in cortical Bdnf expression: Bdnf1 expression was highest at ZT6 and Bdnf4 lowest at ZT12. Such variation was not observed in the basal forebrain. Also Bdnf p1 and p9 methylation levels differed only in the cortex, while Bdnf p4 methylation did not vary in either area. Factorial analysis revealed that sleep deprivation significantly induced Bdnf1 and Bdnf4 with the similar pattern for Bdnf9a in both basal forebrain and cortex; 12 h of sleep deprivation decreased 5mC levels at the cortical Bdnf p4 and p9. Regression analysis between the 5mC promoter levels and the corresponding Bdnf transcript expression revealed significant negative correlations for the basal forebrain Bdnf1 and cortical Bdnf9a transcripts in only non-deprived rats, while these correlations were lost after sleep deprivation. Our results suggest that Bdnf transcription during the light phase of undisturbed sleep-wake cycle but not after SD is regulated at least partially by brain site-specific DNA methylation.',
'date' => '2014-09-15',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25223586',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 89 => array(
'id' => '2109',
'name' => 'Transient accumulation of 5-carboxylcytosine indicates involvement of active demethylation in lineage specification of neural stem cells.',
'authors' => 'Wheldon LM, Abakir A, Ferjentsik Z, Dudnakova T, Strohbuecker S, Christie D, Dai N, Guan S, Foster JM, Corrêa IR, Loose M, Dixon JE, Sottile V, Johnson AD, Ruzov A',
'description' => '5-Methylcytosine (5mC) is an epigenetic modification involved in regulation of gene activity during differentiation. Tet dioxygenases oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). Both 5fC and 5caC can be excised from DNA by thymine-DNA glycosylase (TDG) followed by regeneration of unmodified cytosine via the base excision repair pathway. Despite evidence that this mechanism is operative in embryonic stem cells, the role of TDG-dependent demethylation in differentiation and development is currently unclear. Here, we demonstrate that widespread oxidation of 5hmC to 5caC occurs in postimplantation mouse embryos. We show that 5fC and 5caC are transiently accumulated during lineage specification of neural stem cells (NSCs) in culture and in vivo. Moreover, 5caC is enriched at the cell-type-specific promoters during differentiation of NSCs, and TDG knockdown leads to increased 5fC/5caC levels in differentiating NSCs. Our data suggest that active demethylation contributes to epigenetic reprogramming determining lineage specification in embryonic brain.',
'date' => '2014-06-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24882006',
'doi' => '',
'modified' => '2015-07-24 15:39:03',
'created' => '2015-07-24 15:39:03',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 90 => array(
'id' => '2448',
'name' => 'Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring.',
'authors' => 'Itzhak Y, Ergui I, Young JI',
'description' => 'The high rate of methamphetamine (METH) abuse among young adults and women of childbearing age makes it imperative to determine the long-term effects of METH exposure on the offspring. We hypothesized that parental METH exposure modulates offspring behavior by disrupting epigenetic programming of gene expression in the brain. To simulate the human pattern of drug use, male and female C57Bl/6J mice were exposed to escalating doses of METH or saline from adolescence through adulthood; following mating, females continue to receive drug or saline through gestational day 17. F1 METH male offspring showed enhanced response to cocaine-conditioned reward and hyperlocomotion. Both F1 METH male and female offspring had reduced response to conditioned fear. Cross-fostering experiments have shown that certain behavioral phenotypes were modulated by maternal care of either METH or saline dams. Analysis of offspring hippocampal DNA methylation showed differentially methylated regions as a result of both METH in utero exposure and maternal care. Our results suggest that behavioral phenotypes and epigenotypes of offspring that were exposed to METH in utero are vulnerable to (a) METH exposure during embryonic development, a period when wide epigenetic reprogramming occurs, and (b) postnatal maternal care.Molecular Psychiatry advance online publication, 18 February 2014; doi:10.1038/mp.2014.7.',
'date' => '2014-02-18',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24535458',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 91 => array(
'id' => '1843',
'name' => 'Alterations of epigenetic signatures in hepatocyte nuclear factor 4α deficient mouse liver determined by improved ChIP-qPCR and (h)MeDIP-qPCR assays.',
'authors' => 'Zhang Q, Lei X, Lu H',
'description' => 'Hepatocyte nuclear factor 4α (HNF4α) is a liver-enriched transcription factor essential for liver development and function. In hepatocytes, HNF4α regulates a large number of genes important for nutrient/xenobiotic metabolism and cell differentiation and proliferation. Currently, little is known about the epigenetic mechanism of gene regulation by HNF4α. In this study, the global and specific alterations at the selected gene loci of representative histone modifications and DNA methylations were investigated in Hnf4a-deficient female mouse livers using the improved MeDIP-, hMeDIP- and ChIP-qPCR assay. Hnf4a deficiency significantly increased hepatic total IPed DNA fragments for histone H3 lysine-4 dimethylation (H3K4me2), H3K4me3, H3K9me2, H3K27me3 and H3K4 acetylation, but not for H3K9me3, 5-methylcytosine,or 5-hydroxymethylcytosine. At specific gene loci, the relative enrichments of histone and DNA modifications were changed to different degree in Hnf4a-deficient mouse liver. Among the epigenetic signatures investigated, changes in H3K4me3 correlated the best with mRNA expression. Additionally, Hnf4a-deficient livers had increased mRNA expression of histone H1.2 and H3.3 as well as epigenetic modifiers Dnmt1, Tet3, Setd7, Kmt2c, Ehmt2, and Ezh2. In conclusion, the present study provides convenient improved (h)MeDIP- and ChIP-qPCR assays for epigenetic study. Hnf4a deficiency in young-adult mouse liver markedly alters histone methylation and acetylation, with fewer effects on DNA methylation and 5-hydroxymethylation. The underlying mechanism may be the induction of epigenetic enzymes responsible for the addition/removal of the epigenetic signatures, and/or the loss of HNF4α per se as a key coordinator for epigenetic modifiers.',
'date' => '2014-01-10',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24427299',
'doi' => '',
'modified' => '2015-07-24 15:39:01',
'created' => '2015-07-24 15:39:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 92 => array(
'id' => '1773',
'name' => 'Peroxisome proliferator-activated receptor γ regulates genes involved in insulin/insulin-like growth factor signaling and lipid metabolism during adipogenesis through functionally distinct enhancer classes.',
'authors' => 'Oger F, Dubois-Chevalier J, Gheeraert C, Avner S, Durand E, Froguel P, Salbert G, Staels B, Lefebvre P, Eeckhoute J',
'description' => 'The nuclear receptor peroxisome proliferator-activated receptor (PPAR) is a transcription factor whose expression is induced during adipogenesis and that is required for the acquisition and control of mature adipocyte functions. Indeed, PPAR induces the expression of genes involved in lipid synthesis and storage through enhancers activated during adipocyte differentiation. Here, we show that PPAR also binds to enhancers already active in preadipocytes as evidenced by an active chromatin state including lower DNA methylation levels despite higher CpG content. These constitutive enhancers are linked to genes involved in the insulin/insulin-like growth factor signaling pathway that are transcriptionally induced during adipogenesis but to a lower extent than lipid metabolism genes, because of stronger basal expression levels in preadipocytes. This is consistent with the sequential involvement of hormonal sensitivity and lipid handling during adipocyte maturation and correlates with the chromatin structure dynamics at constitutive and activated enhancers. Interestingly, constitutive enhancers are evolutionary conserved and can be activated in other tissues, in contrast to enhancers controlling lipid handling genes whose activation is more restricted to adipocytes. Thus, PPAR utilizes both broadly active and cell type-specific enhancers to modulate the dynamic range of activation of genes involved in the adipogenic process.',
'date' => '2014-01-10',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24288131',
'doi' => '',
'modified' => '2015-07-24 15:39:01',
'created' => '2015-07-24 15:39:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 93 => array(
'id' => '1572',
'name' => 'Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model.',
'authors' => 'Yafune A, Kawai M, Itahashi M, Kimura M, Nakane F, Mitsumori K, Shibutani M',
'description' => 'Disruptive epigenetic gene control has been shown to be involved in carcinogenesis. To identify key molecules in piperonyl butoxide (PBO)-induced hepatocarcinogenesis, we searched hypermethylated genes using CpG island (CGI) microarrays in non-neoplastic liver cells as a source of proliferative lesions at 25 weeks after tumor promotion with PBO using mice. We further performed methylation-specific polymerase chain reaction (PCR), real-time reverse transcription PCR, and immunohistochemical analysis in PBO-promoted liver tissues. Ebp4.1, Wdr6 and Cmtm6 increased methylation levels in the promoter region by PBO promotion, although Cmtm6 levels were statistically non-significant. These results suggest that PBO promotion may cause altered epigenetic gene regulation in non-neoplastic liver cells surrounding proliferative lesions to allow the facilitation of hepatocarcinogenesis. Both Wdr6 and Cmtm6 showed decreased expression in non-neoplastic liver cells in contrast to positive immunoreactivity in the majority of proliferative lesions produced by PBO promotion. These results suggest that both Wdr6 and Cmtm6 were spared from epigenetic gene modification in proliferative lesions by PBO promotion in contrast to the hypermethylation-mediated downregulation in surrounding liver cells. Considering the effective detection of proliferative lesions, these molecules could be used as detection markers of hepatocellular proliferative lesions and played an important role in hepatocarcinogenesis.',
'date' => '2013-10-09',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23968726',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 94 => array(
'id' => '1569',
'name' => 'Genome-wide screening identifies Plasmodium chabaudi-induced modifications of DNA methylation status of Tlr1 and Tlr6 gene promoters in liver, but not spleen, of female C57BL/6 mice.',
'authors' => 'Al-Quraishy S, Dkhil MA, Abdel-Baki AA, Delic D, Santourlidis S, Wunderlich F',
'description' => '<p>Epigenetic reprogramming of host genes via DNA methylation is increasingly recognized as critical for the outcome of diverse infectious diseases, but information for malaria is not yet available. Here, we investigate the effect of blood-stage malaria of Plasmodium chabaudi on the DNA methylation status of host gene promoters on a genome-wide scale using methylated DNA immunoprecipitation and Nimblegen microarrays containing 2,000 bp oligonucleotide features that were split into -1,500 to -500 bp Ups promoters and -500 to +500 bp Cor promoters, relative to the transcription site, for evaluation of differential DNA methylation. Gene expression was analyzed by Agilent and Affymetrix microarray technology. Challenging of female C57BL/6 mice with 10(6) P. chabaudi-infected erythrocytes resulted in a self-healing outcome of infections with peak parasitemia on day 8 p.i. These infections induced organ-specific modifications of DNA methylation of gene promoters. Among the 17,354 features on Nimblegen arrays, only seven gene promoters were identified to be hypermethylated in the spleen, whereas the liver exhibited 109 hyper- and 67 hypomethylated promoters at peak parasitemia in comparison with non-infected mice. Among the identified genes with differentially methylated Cor-promoters, only the 7 genes Pigr, Ncf1, Klkb1, Emr1, Ndufb11, and Tlr6 in the liver and Apol6 in the spleen were detected to have significantly changed their expression. Remarkably, the Cor promoter of the toll-like receptor Tlr6 became hypomethylated and Tlr6 expression increased by 3.4-fold during infection. Concomitantly, the Ups promoter of the Tlr1 was hypermethylated, but Tlr1 expression also increased by 11.3-fold. TLR6 and TLR1 are known as auxillary receptors to form heterodimers with TLR2 in plasma membranes of macrophages, which recognize different pathogen-associated molecular patterns (PAMPs), as, e.g., intact 3-acyl and sn-2-lyso-acyl glycosylphosphatidylinositols of P. falciparum, respectively. Our data suggest therefore that malaria-induced epigenetic fine-tuning of Tlr6 and Tlr1 through DNA methylation of their gene promoters in the liver is critically important for initial recognition of PAMPs and, thus, for the final self-healing outcome of blood-stage infections with P. chabaudi malaria.</p>',
'date' => '2013-08-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23949311',
'doi' => '',
'modified' => '2017-10-10 10:37:58',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 95 => array(
'id' => '1466',
'name' => 'Characterization of the DNA methylome and its interindividual variation in human peripheral blood monocytes.',
'authors' => 'Shen H, Qiu C, Li J, Tian Q, Deng HW',
'description' => 'AIM: Peripheral blood monocytes (PBMs) play multiple and critical roles in the immune response, and abnormalities in PBMs have been linked to a variety of human disorders. However, the DNA methylation landscape in PBMs is largely unknown. In this study, we characterized epigenome-wide DNA methylation profiles in purified PBMs. MATERIALS & METHODS: PBMs were isolated from freshly collected peripheral blood from 18 unrelated healthy postmenopausal Caucasian females. Epigenome-wide DNA methylation profiles (the methylome) were characterized by using methylated DNA immunoprecipitation combined with high-throughput sequencing. RESULTS: Distinct patterns were revealed at different genomic features. For instance, promoters were commonly (∼58%) found to be unmethylated; whereas protein coding regions were largely (∼84%) methylated. Although CpG-rich and -poor promoters showed distinct methylation patterns, interestingly, a negative correlation between promoter methylation levels and gene transcription levels was consistently observed across promoters with high to low CpG densities. Importantly, we observed substantial interindividual variation in DNA methylation across the individual PBM methylomes and the pattern of this interindividual variation varied between different genomic features, with highly variable regions enriched for repetitive DNA elements. Furthermore, we observed a modest but significant excess (p < 2.2 × 10(-16)) of genes showing a negative correlation between interindividual promoter methylation and transcription levels. These significant genes were enriched in biological processes that are closely related to PBM functions, suggesting that alteration in DNA methylation is likely to be an important mechanism contributing to the interindividual variation in PBM function, and PBM-related phenotypic and disease-susceptibility variation in humans. CONCLUSION: This study represents a comprehensive analysis of the human PBM methylome and its interindividual variation. Our data provide a valuable resource for future epigenomic and multiomic studies, exploring biological and disease-related regulatory mechanisms in PBMs.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23750642',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 96 => array(
'id' => '1463',
'name' => 'Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation.',
'authors' => 'Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD',
'description' => 'Histone post-translational modifications regulate chromatin structure and function largely through interactions with effector proteins that often contain multiple histone-binding domains. While significant progress has been made characterizing individual effector domains, the role of paired domains and how they function in a combinatorial fashion within chromatin are poorly defined. Here we show that the linked tandem Tudor and plant homeodomain (PHD) of UHRF1 (ubiquitin-like PHD and RING finger domain-containing protein 1) operates as a functional unit in cells, providing a defined combinatorial readout of a heterochromatin signature within a single histone H3 tail that is essential for UHRF1-directed epigenetic inheritance of DNA methylation. These findings provide critical support for the "histone code" hypothesis, demonstrating that multivalent histone engagement plays a key role in driving a fundamental downstream biological event in chromatin.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23752590',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 97 => array(
'id' => '1449',
'name' => 'Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs.',
'authors' => 'Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R',
'description' => 'The presence of 5-methylcytidine (m(5)C) in tRNA and rRNA molecules of a wide variety of organisms was first observed more than 40 years ago. However, detection of this modification was limited to specific, abundant, RNA species, due to the usage of low-throughput methods. To obtain a high resolution, systematic, and comprehensive transcriptome-wide overview of m(5)C across the three domains of life, we used bisulfite treatment on total RNA from both gram positive (B. subtilis) and gram negative (E. coli) bacteria, an archaeon (S. solfataricus) and a eukaryote (S. cerevisiae), followed by massively parallel sequencing. We were able to recover most previously documented m(5)C sites on rRNA in the four organisms, and identified several novel sites in yeast and archaeal rRNAs. Our analyses also allowed quantification of methylated m(5)C positions in 64 tRNAs in yeast and archaea, revealing stoichiometric differences between the methylation patterns of these organisms. Molecules of tRNAs in which m(5)C was absent were also discovered. Intriguingly, we detected m(5)C sites within archaeal mRNAs, and identified a consensus motif of AUCGANGU that directs methylation in S. solfataricus. Our results, which were validated using m(5)C-specific RNA immunoprecipitation, provide the first evidence for mRNA modifications in archaea, suggesting that this mode of post-transcriptional regulation extends beyond the eukaryotic domain.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23825970',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 98 => array(
'id' => '1403',
'name' => 'Methyl donor supplementation blocks the adverse effects of maternal high fat diet on offspring physiology.',
'authors' => 'Carlin J, George R, Reyes TM',
'description' => 'Maternal consumption of a high fat diet during pregnancy increases the offspring risk for obesity. Using a mouse model, we have previously shown that maternal consumption of a high fat (60%) diet leads to global and gene specific decreases in DNA methylation in the brain of the offspring. The present experiments were designed to attempt to reverse this DNA hypomethylation through supplementation of the maternal diet with methyl donors, and to determine whether methyl donor supplementation could block or attenuate phenotypes associated with maternal consumption of a HF diet. Metabolic and behavioral (fat preference) outcomes were assessed in male and female adult offspring. Expression of the mu-opioid receptor and dopamine transporter mRNA, as well as global DNA methylation were measured in the brain. Supplementation of the maternal diet with methyl donors attenuated the development of some of the adverse effects seen in offspring from dams fed a high fat diet; including weight gain, increased fat preference (males), changes in CNS gene expression and global hypomethylation in the prefrontal cortex. Notable sex differences were observed. These findings identify the importance of balanced methylation status during pregnancy, particularly in the context of a maternal high fat diet, for optimal offspring outcome.',
'date' => '2013-05-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23658839',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 99 => array(
'id' => '1311',
'name' => 'Naive pluripotency is associated with global DNA hypomethylation.',
'authors' => 'Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, Surani MA, Hajkova P',
'description' => 'Naive pluripotent embryonic stem cells (ESCs) and embryonic germ cells (EGCs) are derived from the preimplantation epiblast and primordial germ cells (PGCs), respectively. We investigated whether differences exist between ESCs and EGCs, in view of their distinct developmental origins. PGCs are programmed to undergo global DNA demethylation; however, we find that EGCs and ESCs exhibit equivalent global DNA methylation levels. Inhibition of MEK and Gsk3b by 2i conditions leads to pronounced reduction in DNA methylation in both cell types. This is driven by Prdm14 and is associated with downregulation of Dnmt3a and Dnmt3b. However, genomic imprints are maintained in 2i, and we report derivation of EGCs with intact genomic imprints. Collectively, our findings establish that culture in 2i instills a naive pluripotent state with a distinctive epigenetic configuration that parallels molecular features observed in both the preimplantation epiblast and nascent PGCs.',
'date' => '2013-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23416945',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 100 => array(
'id' => '1290',
'name' => 'DNA methylation analysis in the intestinal epithelium-effect of cell separation on gene expression and methylation profile.',
'authors' => 'Jenke AC, Postberg J, Raine T, Nayak KM, Molitor M, Wirth S, Kaser A, Parkes M, Heuschkel RB, Orth V, Zilbauer M',
'description' => 'BACKGROUND: Epigenetic signatures are highly cell type specific. Separation of distinct cell populations is therefore desirable for all epigenetic studies. However, to date little information is available on whether separation protocols might influence epigenetic and/or gene expression signatures and hence might be less beneficial. We investigated the influence of two frequently used protocols to isolate intestinal epithelium cells (IECs) from 6 healthy individuals. MATERIALS AND METHODS: Epithelial cells were isolated from small bowel (i.e. terminal ileum) biopsies using EDTA/DTT and enzymatic release followed by magnetic bead sorting via EPCAM labeled microbeads. Effects on gene/mRNA expression were analyzed using a real time PCR based expression array. DNA methylation was assessed by pyrosequencing of bisulfite converted DNA and methylated DNA immunoprecipitation (MeDIP). RESULTS: While cell purity was >95% using both cell separation approaches, gene expression analysis revealed significantly higher mRNA levels of several inflammatory genes in EDTA/DTT when compared to enzymatically released cells. In contrast, DNA methylation of selected genes was less variable and only revealed subtle differences. Comparison of DNA methylation of the epithelial cell marker EPCAM in unseparated whole biopsy samples with separated epithelium (i.e. EPCAM positive and negative fraction) demonstrated significant differences in DNA methylation between all three tissue fractions indicating cell type specific methylation patterns can be masked in unseparated tissue samples. CONCLUSIONS: Taken together, our data highlight the importance of considering the potential effect of cell separation on gene expression as well as DNA methylation signatures. The decision to separate tissue samples will therefore depend on study design and specific separation protocols.',
'date' => '2013-02-08',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23409010',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 101 => array(
'id' => '1065',
'name' => 'Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation.',
'authors' => 'Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD',
'description' => 'A fundamental challenge in mammalian biology has been the elucidation of mechanisms linking DNA methylation and histone post-translational modifications. Human UHRF1 (ubiquitin-like PHD and RING finger domain-containing 1) has multiple domains that bind chromatin, and it is implicated genetically in the maintenance of DNA methylation. However, molecular mechanisms underlying DNA methylation regulation by UHRF1 are poorly defined. Here we show that UHRF1 association with methylated histone H3 Lys9 (H3K9) is required for DNA methylation maintenance. We further show that UHRF1 association with H3K9 methylation is insensitive to adjacent H3 S10 phosphorylation-a known mitotic 'phospho-methyl switch'. Notably, we demonstrate that UHRF1 mitotic chromatin association is necessary for DNA methylation maintenance through regulation of the stability of DNA methyltransferase-1. Collectively, our results define a previously unknown link between H3K9 methylation and the faithful epigenetic inheritance of DNA methylation, establishing a notable mitotic role for UHRF1 in this process.',
'date' => '2012-09-30',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23022729',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 102 => array(
'id' => '960',
'name' => 'Histone acetylation and DNA demethylation of T-cells result in an anaplastic large cell lymphoma-like phenotype.',
'authors' => 'Joosten M, Seitz V, Zimmermann K, Sommerfeld A, Berg E, Lenze D, Leser U, Stein H, Hummel M',
'description' => 'Background. A characteristic feature of anaplastic large cell lymphoma is the significant repression of the T-cell expression program despite its T-cell origin. The reasons for this down-regulation of T-cell phenotype are still unknown. Design and Methods. To elucidate whether epigenetic mechanisms are responsible for the loss of the T-cell phenotype, we treated anaplastic large cell lymphoma and T-cell lymphoma/leukemia cell lines (n=4, each) with epigenetic modifiers to evoke DNA demethylation and histone acetylation. Global gene expression data from treated and untreated cell lines were generated and selected differentially expressed genes were evaluated by real-time RT-PCR and Western Blot analysis. Additionally, histone H3 lysine 27 trimethylation was analyzed by chromatin immunoprecipitation. Results. Combined DNA demethylation and histone acetylation of anaplastic large cell lymphoma cells was not able to reconstitute their T-cell phenotype. Instead, the same treatment induced in T-cells (i) an up-regulation of anaplastic large cell lymphoma-characteristic genes (e.g. ID2, LGALS1, c-JUN) and (ii) an almost complete extinction of their T-cell phenotype including CD3, LCK and ZAP70. In addition, a suppressive trimethylation of histone H3 lysine 27 of important T-cell transcription factor genes (GATA3, LEF1, TCF1) was present in anaplastic large cell lymphoma cells, which is in line with their absence in primary tumour specimens as demonstrated by immunohistochemistry. Conclusions. Our data suggest that epigenetically activated suppressors (e.g. ID2) contribute to the down-regulation of the T-cell expression program in anaplastic large cell lymphoma, which is maintained by trimethylation of histone H3 lysine 27.',
'date' => '2012-08-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22899583',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 103 => array(
'id' => '389',
'name' => 'Growth Arrest and DNA-Damage-Inducible, Beta (GADD45b)-Mediated DNA Demethylation in Major Psychosis.',
'authors' => 'Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A',
'description' => 'Aberrant neocortical DNA methylation has been suggested to be a pathophysiological contributor to psychotic disorders. Recently, a growth arrest and DNA-damage-inducible, beta (GADD45b) protein-coordinated DNA demethylation pathway, utilizing cytidine deaminases and thymidine glycosylases, has been identified in the brain. We measured expression of several members of this pathway in parietal cortical samples from the Stanley Foundation Neuropathology Consortium (SFNC) cohort. We find an increase in GADD45b mRNA and protein in patients with psychosis. In immunohistochemistry experiments using samples from the Harvard Brain Tissue Resource Center, we report an increased number of GADD45b-stained cells in prefrontal cortical layers II, III, and V in psychotic patients. Brain-derived neurotrophic factor IX (BDNF IXabcd) was selected as a readout gene to determine the effects of GADD45b expression and promoter binding. We find that there is less GADD45b binding to the BDNF IXabcd promoter in psychotic subjects. Further, there is reduced BDNF IXabcd mRNA expression, and an increase in 5-methylcytosine and 5-hydroxymethylcytosine at its promoter. On the basis of these results, we conclude that GADD45b may be increased in psychosis compensatory to its inability to access gene promoter regions.',
'date' => '2012-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22048458',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 104 => array(
'id' => '409',
'name' => 'Epigenetic silencing mediated through activated PI3K/AKT signaling in breast cancer.',
'authors' => 'Zuo T, Liu TM, Lan X, Weng YI, Shen R, Gu F, Huang YW, Liyanarachchi S, Deatherage DE, Hsu PY, Taslim C, Ramaswamy B, Shapiro CL, Lin HJ, Cheng AS, Jin VX, Huang TH',
'description' => 'Trimethylation of histone 3 lysine 27 (H3K27me3) is a critical epigenetic mark for the maintenance of gene silencing. Additional accumulation of DNA methylation in target loci is thought to cooperatively support this epigenetic silencing during tumorigenesis. However, molecular mechanisms underlying the complex interplay between the two marks remain to be explored. Here we show that activation of PI3K/AKT signaling can be a trigger of this epigenetic processing at many downstream target genes. We also find that DNA methylation can be acquired at the same loci in cancer cells, thereby reinforcing permanent repression in those losing the H3K27me3 mark. Because of a link between PI3K/AKT signaling and epigenetic alterations, we conducted epigenetic therapies in conjunction with the signaling-targeted treatment. These combined treatments synergistically relieve gene silencing and suppress cancer cell growth in vitro and in xenografts. The new finding has important implications for improving targeted cancer therapies in the future.',
'date' => '2011-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/21216892',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 105 => array(
'id' => '412',
'name' => 'Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping.',
'authors' => 'Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, Weng YI, Deatherage DE, Chen Z, Pereira JS, Lopez R, Russo J, Wang Q, Lamartiniere CA, Nephew KP, Huang TH',
'description' => 'The current concept of epigenetic repression is based on one repressor unit corresponding to one silent gene. This notion, however, cannot adequately explain concurrent silencing of multiple loci observed in large chromosome regions. The long-range epigenetic silencing (LRES) can be a frequent occurrence throughout the human genome. To comprehensively characterize the influence of estrogen signaling on LRES, we analyzed transcriptome, methylome, and estrogen receptor alpha (ESR1)-binding datasets from normal breast epithelia and breast cancer cells. This "omics" approach uncovered 11 large repressive zones (range, 0.35 approximately 5.98 megabases), including a 14-gene cluster located on 16p11.2. In normal cells, estrogen signaling induced transient formation of multiple DNA loops in the 16p11.2 region by bringing 14 distant loci to focal ESR1-docking sites for coordinate repression. However, the plasticity of this free DNA movement was reduced in breast cancer cells. Together with the acquisition of DNA methylation and repressive chromatin modifications at the 16p11.2 loci, an inflexible DNA scaffold may be a novel determinant used by breast cancer cells to reinforce estrogen-mediated repression.',
'date' => '2010-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/20442245',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 106 => array(
'id' => '976',
'name' => 'Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells.',
'authors' => 'Borgel J, Guibert S, Weber M.',
'description' => 'Methylated DNA immunoprecipitation (MeDIP) is an immunocapturing approach for unbiased enrichment of DNA that is methylated on cytosines. The principle is that genomic DNA is randomly sheared by sonication and immunoprecipitated with an antibody that specifically recognizes 5-methylcytidine (5mC), which can be combined with PCR or high-throughput analysis (microarrays, deep sequencing). The MeDIP technique has been originally used to generate DNA methylation profiles on a genome scale in mammals and plants. Here we provide an optimized version of the MeDIP protocol suitable for low amounts of DNA, which can be used to study DNA methylation in cellular populations available in small quantities.',
'date' => '0000-00-00',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22907495',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '120',
'name' => '5-mC Antibody - clone 33D3 SDS US en',
'language' => 'en',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-US-en-GHS_2_0.pdf',
'countries' => 'US',
'modified' => '2020-03-13 15:47:06',
'created' => '2020-03-13 15:47:06',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '118',
'name' => '5-mC Antibody - clone 33D3 SDS GB en',
'language' => 'en',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-GB-en-GHS_2_0.pdf',
'countries' => 'GB',
'modified' => '2020-03-13 15:44:22',
'created' => '2020-03-13 15:44:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '116',
'name' => '5-mC Antibody - clone 33D3 SDS ES es',
'language' => 'es',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-ES-es-GHS_2_0.pdf',
'countries' => 'ES',
'modified' => '2020-03-13 15:43:01',
'created' => '2020-03-13 15:43:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '115',
'name' => '5-mC Antibody - clone 33D3 SDS DE de',
'language' => 'de',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-DE-de-GHS_2_0.pdf',
'countries' => 'DE',
'modified' => '2020-03-13 15:42:22',
'created' => '2020-03-13 15:42:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '119',
'name' => '5-mC Antibody - clone 33D3 SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-JP-ja-GHS_4_0.pdf',
'countries' => 'JP',
'modified' => '2020-03-13 15:44:56',
'created' => '2020-03-13 15:44:56',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '114',
'name' => '5-mC Antibody - clone 33D3 SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-03-13 15:41:00',
'created' => '2020-03-13 15:41:00',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '113',
'name' => '5-mC Antibody - clone 33D3 SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-BE-fr-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-03-13 15:40:12',
'created' => '2020-03-13 15:40:12',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '117',
'name' => '5-mC Antibody - clone 33D3 SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-03-13 15:43:48',
'created' => '2020-03-13 15:43:48',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = true
$other_formats = array(
(int) 0 => array(
'id' => '1980',
'antibody_id' => '630',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>Monoclonal antibody raised in mouse against </span><b>5-mC</b><span><span> </span>(</span><b>5-methylcytosine</b><span>) conjugated to ovalbumine (</span><b>33D3 clone</b><span>).</span></p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15200081-100',
'old_catalog_number' => 'MAb-081-100',
'sf_code' => 'C15200081-D001-000526',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '505',
'price_USD' => '575',
'price_GBP' => '450',
'price_JPY' => '79110',
'price_CNY' => '0',
'price_AUD' => '1438',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'October 27, 2020',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081) | Diagenode',
'meta_keywords' => '5-methylcytosine (5-mC),monoclonal antibody,Methylated DNA Immunoprecipitation',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2023-05-17 10:08:33',
'created' => '2015-06-29 14:08:20'
),
(int) 1 => array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
)
$pro = array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010034</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1885" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1885" id="CartAdd/1885Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1885" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto hMeDIP kit x16 (monoclonal mouse antibody)</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns" data-reveal-id="cartModal-1885" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto hMeDIP kit x16 (monoclonal mouse antibody)</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/magmedip-kit-x48-48-rxns"><img src="/img/product/kits/C02010021-magmedip-qpcr.jpg" alt="MagMeDIP qPCR Kit box" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010021</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1880" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1880" id="CartAdd/1880Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1880" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MagMeDIP Kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MagMeDIP Kit',
'C02010021',
'750',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MagMeDIP Kit',
'C02010021',
'750',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="magmedip-kit-x48-48-rxns" data-reveal-id="cartModal-1880" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MagMeDIP Kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/auto-methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020011</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1888" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1888" id="CartAdd/1888Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1888" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto MethylCap kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1888" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto MethylCap kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1887" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1887" id="CartAdd/1887Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1887" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MethylCap kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MethylCap kit',
'C02020010',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MethylCap kit',
'C02020010',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1887" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MethylCap kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/premium-bisulfite-kit-50-rxns"><img src="/img/grey-logo.jpg" alt="default alt" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02030030</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1892" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1892" id="CartAdd/1892Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1892" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Premium Bisulfite kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Premium Bisulfite kit',
'C02030030',
'240',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Premium Bisulfite kit',
'C02030030',
'240',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="premium-bisulfite-kit-50-rxns" data-reveal-id="cartModal-1892" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Premium Bisulfite kit</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1892',
'antibody_id' => null,
'name' => 'Premium Bisulfite kit',
'description' => '<p style="text-align: center;"><a href="https://www.diagenode.com/files/products/kits/Premium_Bisulfite_kit_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p style="text-align: center;"><strong>Make your Bisulfite conversion now in only 60 minutes !</strong></p>
<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C02030030',
'old_catalog_number' => '',
'sf_code' => 'C02030030-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'premium-bisulfite-kit-50-rxns',
'meta_title' => 'Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Premium Bisulfite kit',
'modified' => '2023-04-20 16:13:50',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '271',
'product_id' => '1979',
'related_id' => '1892'
),
'Image' => array()
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ' <span style="color:#CCC">(MAb-081-500)</span>'
$country_code = 'US'
$other_format = array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
'id' => '1914',
'product_id' => '1979',
'application_id' => '29'
)
)
$slugs = array(
(int) 0 => 'immunofluorescence'
)
$applications = array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'locale' => 'eng'
)
$description = '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>'
$name = 'IF'
$document = array(
'id' => '1118',
'name' => 'Datasheet 5-mC33D3 C15200081-10',
'description' => '<p><span>The 5-methylcytosine antibody (clone 33D3) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span></p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_5-mC33D3_C15200081-10.pdf',
'slug' => 'datasheet-5-mc33d3-c15200081-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-12-17 10:55:09',
'created' => '2020-12-17 10:55:09',
'ProductsDocument' => array(
'id' => '3063',
'product_id' => '1979',
'document_id' => '1118'
)
)
$sds = array(
'id' => '117',
'name' => '5-mC Antibody - clone 33D3 SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-03-13 15:43:48',
'created' => '2020-03-13 15:43:48',
'ProductsSafetySheet' => array(
'id' => '231',
'product_id' => '1979',
'safety_sheet_id' => '117'
)
)
$publication = array(
'id' => '976',
'name' => 'Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells.',
'authors' => 'Borgel J, Guibert S, Weber M.',
'description' => 'Methylated DNA immunoprecipitation (MeDIP) is an immunocapturing approach for unbiased enrichment of DNA that is methylated on cytosines. The principle is that genomic DNA is randomly sheared by sonication and immunoprecipitated with an antibody that specifically recognizes 5-methylcytidine (5mC), which can be combined with PCR or high-throughput analysis (microarrays, deep sequencing). The MeDIP technique has been originally used to generate DNA methylation profiles on a genome scale in mammals and plants. Here we provide an optimized version of the MeDIP protocol suitable for low amounts of DNA, which can be used to study DNA methylation in cellular populations available in small quantities.',
'date' => '0000-00-00',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22907495',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
'id' => '711',
'product_id' => '1979',
'publication_id' => '976'
)
)
$externalLink = ' <a href="http://www.ncbi.nlm.nih.gov/pubmed/22907495" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: header [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'product' => array(
'Product' => array(
'id' => '1979',
'antibody_id' => '631',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (sample size)',
'description' => '<p>Monoclonal antibody raised in mouse against 5-mC (<strong>5-methylcytosine</strong>) conjugated to ovalbumine (<strong>33D3 clone</strong>).</p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15200081-10',
'old_catalog_number' => 'MAb-081-010',
'sf_code' => 'C15200081-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10-ug-5-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:58:36',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '631',
'name' => '5-mC monoclonal antibody 33D3 (sample size)',
'description' => '',
'clonality' => '',
'isotype' => 'IgG1',
'lot' => 'RD-006',
'concentration' => '1.1 µg/µl',
'reactivity' => 'Human, mouse, rat, cow, other (wide range): positive',
'type' => 'Monoclonal. <br /><strong>MeDIP grade.</strong><br /><strong>MeDIP-seq grade</strong>.',
'purity' => 'Protein A purified monoclonal antibody.',
'classification' => '',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>MeDIP/MeDIP-seq <sup>*</sup></td>
<td>0.1 - 2 µg/IP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting **</td>
<td>1:300</td>
<td>Fig 3</td>
</tr>
<tr>
<td>IF</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
</tbody>
</table>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.1-5 µg per IP.</small></p>
<p><small><sup>**</sup> Dot blot was only performed to demonstrate the specificity. This antibody is not recommended for dot blot on biological samples.</small></p>
<p></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2023-05-11 15:21:00',
'created' => '2020-12-17 10:31:21',
'select_label' => '631 - 5-mC monoclonal antibody 33D3 (sample size) (RD-006 - 1.1 µg/µl - Human, mouse, rat, cow, other (wide range): positive - Protein A purified monoclonal antibody. - Mouse)'
),
'Slave' => array(),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
),
(int) 25 => array(
[maximum depth reached]
),
(int) 26 => array(
[maximum depth reached]
),
(int) 27 => array(
[maximum depth reached]
),
(int) 28 => array(
[maximum depth reached]
),
(int) 29 => array(
[maximum depth reached]
),
(int) 30 => array(
[maximum depth reached]
),
(int) 31 => array(
[maximum depth reached]
),
(int) 32 => array(
[maximum depth reached]
),
(int) 33 => array(
[maximum depth reached]
),
(int) 34 => array(
[maximum depth reached]
),
(int) 35 => array(
[maximum depth reached]
),
(int) 36 => array(
[maximum depth reached]
),
(int) 37 => array(
[maximum depth reached]
),
(int) 38 => array(
[maximum depth reached]
),
(int) 39 => array(
[maximum depth reached]
),
(int) 40 => array(
[maximum depth reached]
),
(int) 41 => array(
[maximum depth reached]
),
(int) 42 => array(
[maximum depth reached]
),
(int) 43 => array(
[maximum depth reached]
),
(int) 44 => array(
[maximum depth reached]
),
(int) 45 => array(
[maximum depth reached]
),
(int) 46 => array(
[maximum depth reached]
),
(int) 47 => array(
[maximum depth reached]
),
(int) 48 => array(
[maximum depth reached]
),
(int) 49 => array(
[maximum depth reached]
),
(int) 50 => array(
[maximum depth reached]
),
(int) 51 => array(
[maximum depth reached]
),
(int) 52 => array(
[maximum depth reached]
),
(int) 53 => array(
[maximum depth reached]
),
(int) 54 => array(
[maximum depth reached]
),
(int) 55 => array(
[maximum depth reached]
),
(int) 56 => array(
[maximum depth reached]
),
(int) 57 => array(
[maximum depth reached]
),
(int) 58 => array(
[maximum depth reached]
),
(int) 59 => array(
[maximum depth reached]
),
(int) 60 => array(
[maximum depth reached]
),
(int) 61 => array(
[maximum depth reached]
),
(int) 62 => array(
[maximum depth reached]
),
(int) 63 => array(
[maximum depth reached]
),
(int) 64 => array(
[maximum depth reached]
),
(int) 65 => array(
[maximum depth reached]
),
(int) 66 => array(
[maximum depth reached]
),
(int) 67 => array(
[maximum depth reached]
),
(int) 68 => array(
[maximum depth reached]
),
(int) 69 => array(
[maximum depth reached]
),
(int) 70 => array(
[maximum depth reached]
),
(int) 71 => array(
[maximum depth reached]
),
(int) 72 => array(
[maximum depth reached]
),
(int) 73 => array(
[maximum depth reached]
),
(int) 74 => array(
[maximum depth reached]
),
(int) 75 => array(
[maximum depth reached]
),
(int) 76 => array(
[maximum depth reached]
),
(int) 77 => array(
[maximum depth reached]
),
(int) 78 => array(
[maximum depth reached]
),
(int) 79 => array(
[maximum depth reached]
),
(int) 80 => array(
[maximum depth reached]
),
(int) 81 => array(
[maximum depth reached]
),
(int) 82 => array(
[maximum depth reached]
),
(int) 83 => array(
[maximum depth reached]
),
(int) 84 => array(
[maximum depth reached]
),
(int) 85 => array(
[maximum depth reached]
),
(int) 86 => array(
[maximum depth reached]
),
(int) 87 => array(
[maximum depth reached]
),
(int) 88 => array(
[maximum depth reached]
),
(int) 89 => array(
[maximum depth reached]
),
(int) 90 => array(
[maximum depth reached]
),
(int) 91 => array(
[maximum depth reached]
),
(int) 92 => array(
[maximum depth reached]
),
(int) 93 => array(
[maximum depth reached]
),
(int) 94 => array(
[maximum depth reached]
),
(int) 95 => array(
[maximum depth reached]
),
(int) 96 => array(
[maximum depth reached]
),
(int) 97 => array(
[maximum depth reached]
),
(int) 98 => array(
[maximum depth reached]
),
(int) 99 => array(
[maximum depth reached]
),
(int) 100 => array(
[maximum depth reached]
),
(int) 101 => array(
[maximum depth reached]
),
(int) 102 => array(
[maximum depth reached]
),
(int) 103 => array(
[maximum depth reached]
),
(int) 104 => array(
[maximum depth reached]
),
(int) 105 => array(
[maximum depth reached]
),
(int) 106 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul'
)
$language = 'en'
$meta_keywords = ''
$meta_description = '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.'
$meta_title = '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode'
$product = array(
'Product' => array(
'id' => '1979',
'antibody_id' => '631',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (sample size)',
'description' => '<p>Monoclonal antibody raised in mouse against 5-mC (<strong>5-methylcytosine</strong>) conjugated to ovalbumine (<strong>33D3 clone</strong>).</p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15200081-10',
'old_catalog_number' => 'MAb-081-010',
'sf_code' => 'C15200081-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10-ug-5-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:58:36',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '631',
'name' => '5-mC monoclonal antibody 33D3 (sample size)',
'description' => '',
'clonality' => '',
'isotype' => 'IgG1',
'lot' => 'RD-006',
'concentration' => '1.1 µg/µl',
'reactivity' => 'Human, mouse, rat, cow, other (wide range): positive',
'type' => 'Monoclonal. <br /><strong>MeDIP grade.</strong><br /><strong>MeDIP-seq grade</strong>.',
'purity' => 'Protein A purified monoclonal antibody.',
'classification' => '',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>MeDIP/MeDIP-seq <sup>*</sup></td>
<td>0.1 - 2 µg/IP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting **</td>
<td>1:300</td>
<td>Fig 3</td>
</tr>
<tr>
<td>IF</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
</tbody>
</table>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.1-5 µg per IP.</small></p>
<p><small><sup>**</sup> Dot blot was only performed to demonstrate the specificity. This antibody is not recommended for dot blot on biological samples.</small></p>
<p></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2023-05-11 15:21:00',
'created' => '2020-12-17 10:31:21',
'select_label' => '631 - 5-mC monoclonal antibody 33D3 (sample size) (RD-006 - 1.1 µg/µl - Human, mouse, rat, cow, other (wide range): positive - Protein A purified monoclonal antibody. - Mouse)'
),
'Slave' => array(),
'Group' => array(
'Group' => array(
'id' => '4',
'name' => 'C15200081',
'product_id' => '1980',
'modified' => '2016-02-17 17:32:42',
'created' => '2016-02-17 17:32:42'
),
'Master' => array(
'id' => '1980',
'antibody_id' => '630',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>Monoclonal antibody raised in mouse against </span><b>5-mC</b><span><span> </span>(</span><b>5-methylcytosine</b><span>) conjugated to ovalbumine (</span><b>33D3 clone</b><span>).</span></p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15200081-100',
'old_catalog_number' => 'MAb-081-100',
'sf_code' => 'C15200081-D001-000526',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '505',
'price_USD' => '575',
'price_GBP' => '450',
'price_JPY' => '79110',
'price_CNY' => '0',
'price_AUD' => '1438',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'October 27, 2020',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081) | Diagenode',
'meta_keywords' => '5-methylcytosine (5-mC),monoclonal antibody,Methylated DNA Immunoprecipitation',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2023-05-17 10:08:33',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1885',
'antibody_id' => null,
'name' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'description' => '<p><span>The Auro hMeDIP kit is designed for enrichment of hydroxymethylated DNA from fragmented genomic DNA samples for use in genome-wide methylation analysis. It features</span><span> a highly specific monoclonal antibody against </span><span>5-hydroxymethylcytosine (5-hmC) for the immunoprecipitation of hydroxymethylated DNA</span><span>. It includes control DNA and primers to assess the effiency of the assay. </span><span>Performing hydroxymethylation profiling with the hMeDIP kit is fast, reliable and highly specific.</span></p>',
'label1' => ' Characteristics',
'info1' => '<ul style="list-style-type: disc;">
<li><span>Robust enrichment & immunoprecipitation of hydroxymethylated DNA</span></li>
<li>Highly specific monoclonal antibody against 5-hmC<span> for reliable, reproducible results</span></li>
<li>Including control DNA and primers to <span>monitor the efficiency of the assay</span>
<ul style="list-style-type: circle;">
<li>5-hmC, 5-mC and unmethylated DNA sequences and primer pairs</li>
<li>Mouse primer pairs against Sfi1 targeting hydroxymethylated gene in mouse</li>
</ul>
</li>
</ul>
<ul style="list-style-type: disc;">
<li>Improved single-tube, magnetic bead-based protocol</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C02010034',
'old_catalog_number' => '',
'sf_code' => 'C02010034-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '630',
'price_USD' => '690',
'price_GBP' => '580',
'price_JPY' => '98690',
'price_CNY' => '',
'price_AUD' => '1725',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns',
'meta_title' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'meta_keywords' => '',
'meta_description' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'modified' => '2021-01-18 10:37:19',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1880',
'antibody_id' => null,
'name' => 'MagMeDIP Kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/magmedip-kit-manual-C02010020-21.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p> </p>
<div class="small-12 medium-4 large-4 columns"><center></center><center></center><center></center><center><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank"><img src="https://www.diagenode.com/img/banners/banner-nature-publication-580.png" alt="Click here to read more about MeDIP " caption="false" width="80%" /></a></center></div>
<div class="small-12 medium-8 large-8 columns">
<h3 style="text-align: justify;">Sensitive tumour detection and classification using plasma cell-free DNA methylomes<br /><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank">Read the publication</a></h3>
<h3 class="c-article-title u-h1" data-test="article-title" itemprop="name headline" style="text-align: justify;">Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA<br /><a href="https://www.nature.com/articles/s41596-019-0202-2" target="_blank" title="cfMeDIP-seq Nature Method">Read the method</a></h3>
</div>
<p></p>
<p></p>
<p></p>
<div class="row">
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
<div class="small-12 medium-8 large-8 columns"><br />
<p>Perform <strong>MeDIP</strong> (<strong>Me</strong>thylated <strong>D</strong>NA <strong>I</strong>mmuno<strong>p</strong>recipitation) followed by qPCR or NGS to estimate DNA methylation status of your sample using a highly sensitive 5-methylcytosine antibody. Our MagMeDIP kit contains high quality reagents to get the highest enrichment of methylated DNA with an optimized user-friendly protocol.</p>
</div>
</div>
<h3><span>Features</span></h3>
<ul>
<li>Starting DNA amount: <strong>10 ng – 1 µg</strong></li>
<li>Content: <strong>all reagents included</strong> for DNA extraction, immunoprecipitation (including the 5-mC antibody, spike-in controls and their corresponding qPCR primer pairs) as well as DNA isolation after IP.</li>
<li>Application: <strong>qPCR</strong> and <strong>NGS</strong></li>
<li>Robust method, <strong>superior enrichment</strong>, and easy-to-use protocol</li>
<li><strong>High reproducibility</strong> between replicates and repetitive experiments</li>
<li>Compatible with <strong>all species </strong></li>
</ul>',
'label1' => 'MagMeDIP workflow',
'info1' => '<p>DNA methylation occurs primarily as 5-methylcytosine (5-mC), and the Diagenode MagMeDIP Kit takes advantage of a specific antibody targeting this 5-mC to immunoprecipitate methylated DNA, which can be thereafter directly analyzed by qPCR or Next-Generation Sequencing (NGS).</p>
<h3><span>How it works</span></h3>
<p>In brief, after the cell collection and lysis, the genomic DNA is extracted, sheared, and then denatured. In the next step the antibody directed against 5 methylcytosine and antibody binding beads are used for immunoselection and immunoprecipitation of methylated DNA fragments. Then, the IP’d methylated DNA is isolated and can be used for any subsequent analysis as qPCR, amplification, hybridization on microarrays or next generation sequencing.</p>
<center><img src="https://www.diagenode.com/img/product/kits/MagMeDIP-workflow.png" width="70%" alt="5-methylcytosine" caption="false" /></center>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>',
'label2' => 'MeDIP-qPCR',
'info2' => '<p>The kit MagMeDIP contains all reagents necessary for a complete MeDIP-qPCR workflow. Two MagMeDIP protocols have been validated: for manual processing as well as for automated processing, using the Diagenode’s IP-Star Compact Automated System (please refer to the kit manual).</p>
<ul>
<li><strong>Complete kit</strong> including DNA extraction module, IP antibody and reagents, DNA isolation buffer</li>
<li><strong>Quality control of the IP:</strong> due to methylated and unmethylated DNA spike-in controls and their associated qPCR primers</li>
<li><strong>Easy to use</strong> with user-friendly magnetic beads and rack</li>
<li><strong>Highly validated protocol</strong></li>
<li>Automated protocol supplied</li>
</ul>
<center><img src="https://www.diagenode.com/img/product/kits/fig1-magmedipkit.png" width="85%" alt="Methylated DNA Immunoprecipitation" caption="false" /></center>
<p style="font-size: 0.9em;"><em><strong>Figure 1.</strong> Immunoprecipitation results obtained with Diagenode MagMeDIP Kit</em></p>
<p style="font-size: 0.9em;">MeDIP assays were performed manually using 1 µg or 50 ng gDNA from blood cells with the MagMeDIP kit (Diagenode). The IP was performed with the Methylated and Unmethylated spike-in controls included in the kit, together with the human DNA samples. The DNA was isolated/purified using DIB. Afterwards, qPCR was performed using the primer pairs included in this kit.</p>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>',
'label3' => 'MeDIP-seq',
'info3' => '<p>For DNA methylation analysis on the whole genome, MagMeDIP kit can be coupled with Next-Generation Sequencing. To perform MeDIP-sequencing we recommend the following strategy:</p>
<ul style="list-style-type: circle;">
<li>Choose a library preparation solution which is compatible with the starting amount of DNA you are planning to use (from 10 ng to 1 μg). It can be a home-made solution or a commercial one.</li>
<li>Choose the indexing system that fits your needs considering the following features:</li>
<ul>
<ul>
<ul>
<li>Single-indexing, combinatorial dual-indexing or unique dual-indexing</li>
<li>Number of barcodes</li>
<li>Full-length adaptors containing the barcodes or barcoding at the final amplification step</li>
<li>Presence / absence of Unique Molecular Identifiers (for PCR duplicates removal)</li>
</ul>
</ul>
</ul>
<li>Standard library preparation protocols are compatible with double-stranded DNA only, therefore the first steps of the library preparation (end repair, A-tailing, adaptor ligation and clean-up) will have to be performed on sheared DNA, before the IP.</li>
</ul>
<p style="padding-left: 30px;"><strong>CAUTION:</strong> As the immunoprecipitation step occurs at the middle of the library preparation workflow, single-tube solutions for library preparation are usually not compatible with MeDIP-sequencing.</p>
<ul style="list-style-type: circle;">
<li>For DNA isolation after the IP, we recommend using the <a href="https://www.diagenode.com/en/p/ipure-kit-v2-x24" title="IPure kit v2">IPure kit v2</a> (available separately, Cat. No. C03010014) instead of DNA isolation Buffer.</li>
</ul>
<ul style="list-style-type: circle;">
<li>Perform library amplification after the DNA isolation following the standard protocol of the chosen library preparation solution.</li>
</ul>
<h3><span>MeDIP-seq workflow</span></h3>
<center><img src="https://www.diagenode.com/img/product/kits/MeDIP-seq-workflow.png" width="110%" alt="MagMeDIP qPCR Kit x10 workflow" caption="false" /></center>
<h3><span>Example of results</span></h3>
<center><img src="https://www.diagenode.com/img/product/kits/medip-specificity.png" alt="MagMeDIP qPCR Kit Result" caption="false" width="951" height="488" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 1. qPCR analysis of external spike-in DNA controls (methylated and unmethylated) after IP.</strong> Samples were prepared using 1μg – 100ng -10ng sheared human gDNA with the MagMeDIP kit (Diagenode) and a commercially available library prep kit. DNA isolation after IP has been performed with IPure kit V2 (Diagenode).</p>
<p></p>
<p></p>
<center><img src="https://www.diagenode.com/img/product/kits/medip-saturation-analysis.png" alt=" MagMeDIP kit " caption="false" width="951" height="461" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 2. Saturation analysis.</strong> Clean reads were aligned to the human genome (hg19) using Burrows-Wheeler aligner (BWA) algorithm after which duplicated and unmapped reads were removed resulting in a mapping efficiency >98% for all samples. Quality and validity check of the mapped MeDIP-seq data was performed using MEDIPS R package. Saturation plots show that all sets of reads have sufficient complexity and depth to saturate the coverage profile of the reference genome and that this is reproducible between replicates and repetitive experiments (data shown for 50 ng gDNA input: left panel = replicate a, right panel = replicate b).</p>
<p></p>
<p></p>
<center><img src="https://www.diagenode.com/img/product/kits/medip-libraries-prep.png" alt="MagMeDIP x10 " caption="false" width="951" height="708" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 3. Sequencing profiles of MeDIP-seq libraries prepared from different starting amounts of sheared gDNA on the positive and negative methylated control regions.</strong> MeDIP-seq libraries were prepared from decreasing starting amounts of gDNA (1 μg (green), 50 ng (red), and 10ng (blue)) originating from human blood with the MagMeDIP kit (Diagenode) and a commercially available library prep kit. DNA isolation after IP has been performed with IPure kit V2 (Diagenode). IP and corresponding INPUT samples were sequenced on Illumina NovaSeq SP with 2x50 PE reads. The reads were mapped to the human genome (hg19) with bwa and the alignments were loaded into IGV (the tracks use an identical scale). The top IGV figure shows the TSH2B (also known as H2BC1) gene (marked by blue boxes in the bottom track) and its surroundings. The TSH2B gene is coding for a histone variant that does not occur in blood cells, and it is known to be silenced by methylation. Accordingly, we see a high coverage in the vicinity of this gene. The bottom IGV figure shows the GADPH locus (marked by blue boxes in the bottom track) and its surroundings. The GADPH gene is a highly active transcription region and should not be methylated, resulting in no reads accumulation following MeDIP-seq experiment.</p>
<p></p>
<ul>
<ul>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
</ul>
</ul>',
'format' => '48 rxns (IP)',
'catalog_number' => 'C02010021',
'old_catalog_number' => 'mc-magme-048',
'sf_code' => 'C02010021-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '745',
'price_USD' => '750',
'price_GBP' => '680',
'price_JPY' => '116705',
'price_CNY' => '',
'price_AUD' => '1875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'magmedip-kit-x48-48-rxns',
'meta_title' => 'MagMeDIP Kit for efficient immunoprecipitation of methylated DNA | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Perform Methylated DNA Immunoprecipitation (MeDIP) to estimate DNA methylation status of your sample using highly specific 5-mC antibody. This kit allows the preparation of cfMeDIP-seq libraries.',
'modified' => '2023-07-06 14:15:10',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1887',
'antibody_id' => null,
'name' => 'MethylCap kit',
'description' => '<p>The MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. The procedure has been adapted to both manual process or <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a>. Libraries of captured methylated DNA can be prepared for next-generation sequencing (NGS) by combining MBD technology with the <a href="https://www.diagenode.com/en/p/microplex-lib-prep-kit-v3-48-rxns">MicroPlex Library Preparation Kit v3</a>.</p>',
'label1' => 'Characteristics',
'info1' => '<ul style="list-style-type: circle;">
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>On-day protocol</strong></li>
<li><strong>NGS compatibility</strong></li>
</ul>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong></strong></p>
<p><strong></strong><strong>F</strong><strong>igure 1.</strong> Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).</p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020010',
'old_catalog_number' => 'AF-100-0048',
'sf_code' => 'C02020010-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'methylcap-kit-x48-48-rxns',
'meta_title' => 'MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'MethylCap kit x48',
'modified' => '2021-02-19 17:55:38',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '1892',
'antibody_id' => null,
'name' => 'Premium Bisulfite kit',
'description' => '<p style="text-align: center;"><a href="https://www.diagenode.com/files/products/kits/Premium_Bisulfite_kit_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p style="text-align: center;"><strong>Make your Bisulfite conversion now in only 60 minutes !</strong></p>
<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C02030030',
'old_catalog_number' => '',
'sf_code' => 'C02030030-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'premium-bisulfite-kit-50-rxns',
'meta_title' => 'Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Premium Bisulfite kit',
'modified' => '2023-04-20 16:13:50',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array([maximum depth reached])
)
),
'Application' => array(
(int) 0 => array(
'id' => '28',
'position' => '10',
'parent_id' => '40',
'name' => 'DB',
'description' => '<p>Dot blotting</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'dot-blotting',
'meta_keywords' => 'Dot blotting,Monoclonal & Polyclonal antibody,',
'meta_description' => 'Diagenode offers Monoclonal & Polyclonal antibodies for Dot blotting applications',
'meta_title' => 'Dot blotting Antibodies - Monoclonal & Polyclonal antibody | Diagenode',
'modified' => '2016-01-13 14:40:49',
'created' => '2015-07-08 13:45:05',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '23',
'position' => '40',
'parent_id' => '4',
'name' => 'DNA modifications',
'description' => '<p><span style="font-weight: 400;">T</span><span style="font-weight: 400;">he pattern of <strong>DNA modifications</strong> is critical for genome stability and the control of gene expression in the cell. Methylation of 5-cytosine (5-mC), one of the best-studied epigenetic marks, is carried out by the <strong>DNA methyltransferases</strong> DNMT3A and B and DNMT1. DNMT3A and DNMT3B are responsible for </span><i><span style="font-weight: 400;">de novo</span></i><span style="font-weight: 400;"> DNA methylation, whereas DNMT1 maintains existing methylation. 5-mC undergoes active demethylation which is performed by the <strong>Ten-Eleven Translocation</strong> (TET) familly of DNA hydroxylases. The latter consists of 3 members TET1, 2 and 3. All 3 members catalyze the conversion of <strong>5-methylcytosine</strong> (5-mC) into <strong>5-hydroxymethylcytosine</strong> (5-hmC), and further into <strong>5-formylcytosine</strong> (5-fC) and <strong>5-carboxycytosine</strong> (5-caC). 5-fC and 5-caC can be converted to unmodified cytosine by <strong>Thymine DNA Glycosylase</strong> (TDG). It is not yet clear if 5-hmC, 5-fC and 5-caC have specific functions or are simply intermediates in the demethylation of 5-mC.</span></p>
<p><span style="font-weight: 400;">DNA methylation is generally considered as a repressive mark and is usually associated with gene silencing. It is essential that the balance between DNA methylation and demethylation is precisely maintained. Dysregulation of DNA methylation may lead to many different human diseases and is often observed in cancer cells.</span></p>
<p><span style="font-weight: 400;">Diagenode offers highly validated antibodies against different proteins involved in DNA modifications as well as against the modified bases allowing the study of all steps and intermediates in the DNA methylation/demethylation pathway:</span></p>
<p><img src="https://www.diagenode.com/img/categories/antibodies/dna-methylation.jpg" height="599" width="816" /></p>
<p><strong>Diagenode exclusively sources the original 5-methylcytosine monoclonal antibody (clone 33D3).</strong></p>
<p>Check out the list below to see all proposed antibodies for DNA modifications.</p>
<p>Diagenode’s highly validated antibodies:</p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'dna-methylation-antibody',
'cookies_tag_id' => null,
'meta_keywords' => 'Monoclonal Antibodies,Polyclonal antibodies,DNA methylation,Diagenode',
'meta_description' => 'Diagenode offers Monoclonal and Polyclonal antibodies for DNA Methylation. The pattern of DNA modifications is critical for genome stability and the control of gene expression in the cell. ',
'meta_title' => 'DNA modifications - Monoclonal and Polyclonal Antibodies for DNA Methylation | Diagenode',
'modified' => '2020-08-03 15:25:37',
'created' => '2014-07-31 03:51:53',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '102',
'position' => '1',
'parent_id' => '4',
'name' => 'Sample size antibodies',
'description' => '<h1><strong>Validated epigenetics antibodies</strong> – care for a sample?<br /> </h1>
<p>Diagenode has partnered with leading epigenetics experts and numerous epigenetics consortiums to bring to you a validated and comprehensive collection of epigenetic antibodies. As an expert in epigenetics, we are committed to offering highly-specific antibodies validated for ChIP/ChIP-seq and many other applications. All batch-specific validation data is available on our website.<br /><a href="../categories/antibodies">Read about our expertise in antibody production</a>.</p>
<ul>
<li><strong>Focused</strong> - Diagenode's selection of antibodies is exclusively dedicated for epigenetic research. <a title="See the full collection." href="../categories/all-antibodies">See the full collection.</a></li>
<li><strong>Strict quality standards</strong> with rigorous QC and validation</li>
<li><strong>Classified</strong> based on level of validation for flexibility of application</li>
</ul>
<p>Existing sample sizes are listed below. We will soon expand our collection. Are you looking for a sample size of another antibody? Just <a href="mailto:agnieszka.zelisko@diagenode.com?Subject=Sample%20Size%20Request" target="_top">Contact us</a>.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => true,
'is_antibody' => true,
'slug' => 'sample-size-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => '5-hmC monoclonal antibody,CRISPR/Cas9 polyclonal antibody ,H3K36me3 polyclonal antibody,diagenode',
'meta_description' => 'Diagenode offers sample volume on selected antibodies for researchers to test, validate and provide confidence and flexibility in choosing from our wide range of antibodies ',
'meta_title' => 'Sample-size Antibodies | Diagenode',
'modified' => '2019-07-03 10:57:05',
'created' => '2015-10-27 12:13:34',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '15',
'name' => 'Monoclonal Antibody hMeDIP Kit for DNA Hydroxymethylation Studies',
'description' => '<p><span>There is substantial interest and speculation in the role of the “sixth DNA base,” 5-hydroxymethylcytosine (5-hmC), although its precise function has not yet been elucidated. Since its discovery in neuronal Purkinje, granule and ES cells, studies of this new modified DNA base have been limited by the lack of high-quality, validated tools and technologies that discriminate hydroxy-methylation from methylation in regulating genome expression. Obtaining a specific assay for 5-hmC is particularly important since standard bisulfite sequencing cannot distinguish between these two types of methylation. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Monoclonal_Antibody_hMeDIP_Kit_for_DNA_Hydroxymethylation_Studies_Poster.pdf',
'slug' => 'monoclonal-antibody-hmedip-kit-for-dna-hydroxymethylation-studies-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-09-29 13:42:52',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1118',
'name' => 'Datasheet 5-mC33D3 C15200081-10',
'description' => '<p><span>The 5-methylcytosine antibody (clone 33D3) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span></p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_5-mC33D3_C15200081-10.pdf',
'slug' => 'datasheet-5-mc33d3-c15200081-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-12-17 10:55:09',
'created' => '2020-12-17 10:55:09',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '250',
'name' => 'product/antibodies/antibody.png',
'alt' => 'Mouse IgG',
'modified' => '2020-11-27 07:00:09',
'created' => '2015-07-17 10:12:18',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4843',
'name' => 'Differentiation block in acute myeloid leukemia regulated by intronicsequences of FTO',
'authors' => 'Camera F. et al.',
'description' => '<p>Iroquois transcription factor gene IRX3 is highly expressed in 20–30\% of acute myeloid leukemia (AML) and contributes to the pathognomonic differentiation block. Intron 8 FTO sequences ∼220kB downstream of IRX3 exhibit histone acetylation, DNA methylation, and contacts with the IRX3 promoter, which correlate with IRX3 expression. Deletion of these intronic elements confirms a role in positively regulating IRX3. RNAseq revealed long non-coding (lnc) transcripts arising from this locus. FTO-lncAML knockdown (KD) induced differentiation of AML cells, loss of clonogenic activity, and reduced FTO intron 8:IRX3 promoter contacts. While both FTO-lncAML KD and IRX3 KD induced differentiation, FTO-lncAML but not IRX3 KD led to HOXA downregulation suggesting transcript activity in trans. FTO-lncAMLhigh AML samples expressed higher levels of HOXA and lower levels of differentiation genes. Thus, a regulatory module in FTO intron 8 consisting of clustered enhancer elements and a long non-coding RNA is active in human AML, impeding myeloid differentiation.</p>',
'date' => '2023-08-01',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2589004223013962',
'doi' => '10.1016/j.isci.2023.107319',
'modified' => '2023-08-01 14:14:01',
'created' => '2023-08-01 15:59:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4796',
'name' => 'Nicotinamide N-methyltransferase sustains a core epigenetic programthat promotes metastatic colonization in breast cancer.',
'authors' => 'Couto J.P. et al.',
'description' => '<p><span>Metastatic colonization of distant organs accounts for over 90% of deaths related to solid cancers, yet the molecular determinants of metastasis remain poorly understood. Here, we unveil a mechanism of colonization in the aggressive basal-like subtype of breast cancer that is driven by the NAD</span><sup>+</sup><span><span> </span>metabolic enzyme nicotinamide N-methyltransferase (NNMT). We demonstrate that NNMT imprints a basal genetic program into cancer cells, enhancing their plasticity. In line, NNMT expression is associated with poor clinical outcomes in patients with breast cancer. Accordingly, ablation of NNMT dramatically suppresses metastasis formation in pre-clinical mouse models. Mechanistically, NNMT depletion results in a methyl overflow that increases histone H3K9 trimethylation (H3K9me3) and DNA methylation at the promoters of PR/SET Domain-5 (PRDM5) and extracellular matrix-related genes. PRDM5 emerged in this study as a pro-metastatic gene acting via induction of cancer-cell intrinsic transcription of collagens. Depletion of PRDM5 in tumor cells decreases COL1A1 deposition and impairs metastatic colonization of the lungs. These findings reveal a critical activity of the NNMT-PRDM5-COL1A1 axis for cancer cell plasticity and metastasis in basal-like breast cancer.</span></p>',
'date' => '2023-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37259596',
'doi' => '10.15252/embj.2022112559',
'modified' => '2023-06-15 08:35:19',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4787',
'name' => 'The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.',
'authors' => 'Garcia-Gomez E. et al.',
'description' => '<p>Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the disease (control group). In addition, eight of the PCOS patients underwent intervention with metformin (1500 mg/day) and a carbohydrate-controlled diet (type and quantity) for three months. Clinical and metabolic parameters were determined, and RT-qPCR and MeDIP-qPCR were used to evaluate gene expression and DNA methylation levels, respectively. Decreased expression levels of , , and genes and increased DNA methylation levels of the promoter were found in the endometrium of PCOS patients compared to controls. After metformin and nutritional intervention, some metabolic and clinical variables improved in PCOS patients. This intervention was associated with increased expression of , , and genes and reduced DNA methylation levels of the promoter in the endometrium of PCOS women. Our preliminary findings suggest that metformin and a carbohydrate-controlled diet improve endometrial function in PCOS patients, partly by modulating DNA methylation of the gene promoter and the expression of genes implicated in endometrial receptivity and insulin signaling.</p>',
'date' => '2023-04-01',
'pmid' => 'https://doi.org/10.3390%2Fijms24076857',
'doi' => '10.3390/ijms24076857',
'modified' => '2023-06-12 08:58:33',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4777',
'name' => 'Epigenetic modifier alpha-ketoglutarate modulates aberrant gene bodymethylation and hydroxymethylation marks in diabetic heart.',
'authors' => 'Dhat R. et al.',
'description' => '<p>BACKGROUND: Diabetic cardiomyopathy (DCM) is a leading cause of death in diabetic patients. Hyperglycemic myocardial microenvironment significantly alters chromatin architecture and the transcriptome, resulting in aberrant activation of signaling pathways in a diabetic heart. Epigenetic marks play vital roles in transcriptional reprogramming during the development of DCM. The current study is aimed to profile genome-wide DNA (hydroxy)methylation patterns in the hearts of control and streptozotocin (STZ)-induced diabetic rats and decipher the effect of modulation of DNA methylation by alpha-ketoglutarate (AKG), a TET enzyme cofactor, on the progression of DCM. METHODS: Diabetes was induced in male adult Wistar rats with an intraperitoneal injection of STZ. Diabetic and vehicle control animals were randomly divided into groups with/without AKG treatment. Cardiac function was monitored by performing cardiac catheterization. Global methylation (5mC) and hydroxymethylation (5hmC) patterns were mapped in the Left ventricular tissue of control and diabetic rats with the help of an enrichment-based (h)MEDIP-sequencing technique by using antibodies specific for 5mC and 5hmC. Sequencing data were validated by performing (h)MEDIP-qPCR analysis at the gene-specific level, and gene expression was analyzed by qPCR. The mRNA and protein expression of enzymes involved in the DNA methylation and demethylation cycle were analyzed by qPCR and western blotting. Global 5mC and 5hmC levels were also assessed in high glucose-treated DNMT3B knockdown H9c2 cells. RESULTS: We found the increased expression of DNMT3B, MBD2, and MeCP2 with a concomitant accumulation of 5mC and 5hmC, specifically in gene body regions of diabetic rat hearts compared to the control. Calcium signaling was the most significantly affected pathway by cytosine modifications in the diabetic heart. Additionally, hypermethylated gene body regions were associated with Rap1, apelin, and phosphatidyl inositol signaling, while metabolic pathways were most affected by hyperhydroxymethylation. AKG supplementation in diabetic rats reversed aberrant methylation patterns and restored cardiac function. Hyperglycemia also increased 5mC and 5hmC levels in H9c2 cells, which was normalized by DNMT3B knockdown or AKG supplementation. CONCLUSION: This study demonstrates that reverting hyperglycemic damage to cardiac tissue might be possible by erasing adverse epigenetic signatures by supplementing epigenetic modulators such as AKG along with an existing antidiabetic treatment regimen.</p>',
'date' => '2023-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37101286',
'doi' => '10.1186/s13072-023-00489-4',
'modified' => '2023-06-12 09:20:54',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '4780',
'name' => 'Integrated analysis from multicentre studies identities RNAmethylation- related lncRNA risk stratification systems for glioma',
'authors' => 'Huang Fanxuan and Wang Xinyu and Zhong Junzhe and Chen Hao and Song Dan and Xu Tianye and Tian Kaifu and Sun Penggang and Sun Nan and Ma Wenbin and Liu Yuxiang andYu Daohan and Meng Xiangqi and Jiang Chuanlu and Xuan Hanwen and Qian Da an',
'description' => '<p>Gastric cancer (GC) is the fourth leading cause of cancer death worldwide. Due to the lack of effective chemotherapy methods for advanced gastric cancer and poor prognosis, the emergence of immunotherapy has brought new hope to gastric cancer. Further research is needed to improve the response rate to immunotherapy and identify the populations with potential benefits of immunotherapy. It is unclear whether m7G-related lncRNAs influence tumour immunity and the prognosis of immunotherapy.</p>',
'date' => '2023-03-02',
'pmid' => 'https://doi.org/10.21203%2Frs.3.rs-2815231%2Fv1',
'doi' => '10.21203/rs.3.rs-2815231/v1',
'modified' => '2023-06-13 09:25:12',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '4773',
'name' => 'The RNA m5C Methylase NSUN2 Modulates Corneal EpithelialWound Healing.',
'authors' => 'Luo G. et al.',
'description' => '<p>PURPOSE: The emerging epitranscriptomics offers insights into the physiopathological roles of various RNA modifications. The RNA methylase NOP2/Sun domain family member 2 (NSUN2) catalyzes 5-methylcytosine (m5C) modification of mRNAs. However, the role of NSUN2 in corneal epithelial wound healing (CEWH) remains unknown. Here we describe the functional mechanisms of NSUN2 in mediating CEWH. METHODS: RT-qPCR, Western blot, dot blot, and ELISA were used to determine the NSUN2 expression and overall RNA m5C level during CEWH. NSUN2 silencing or overexpression was performed to explore its involvement in CEWH both in vivo and in vitro. Multi-omics was integrated to reveal the downstream target of NSUN2. MeRIP-qPCR, RIP-qPCR, and luciferase assay, as well as in vivo and in vitro functional assays, clarified the molecular mechanism of NSUN2 in CEWH. RESULTS: The NSUN2 expression and RNA m5C level increased significantly during CEWH. NSUN2 knockdown significantly delayed CEWH in vivo and inhibited human corneal epithelial cells (HCEC) proliferation and migration in vitro, whereas NSUN2 overexpression prominently enhanced HCEC proliferation and migration. Mechanistically, we found that NSUN2 increased ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) translation through the binding of RNA m5C reader Aly/REF export factor. Accordingly, UHRF1 knockdown significantly delayed CEWH in vivo and inhibited HCEC proliferation and migration in vitro. Furthermore, UHRF1 overexpression effectively rescued the inhibitory effect of NSUN2 silencing on HCEC proliferation and migration. CONCLUSIONS: NSUN2-mediated m5C modification of UHRF1 mRNA modulates CEWH. This finding highlights the critical importance of this novel epitranscriptomic mechanism in control of CEWH.</p>',
'date' => '2023-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36862118',
'doi' => '10.1167/iovs.64.3.5',
'modified' => '2023-04-17 09:48:55',
'created' => '2023-04-14 13:41:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '4674',
'name' => 'Methylation and expression of glucocorticoid receptor exon-1 variants andFKBP5 in teenage suicide-completers.',
'authors' => 'Rizavi H. et al.',
'description' => '<p>A dysregulated hypothalamic-pituitary-adrenal (HPA) axis has repeatedly been demonstrated to play a fundamental role in psychiatric disorders and suicide, yet the mechanisms underlying this dysregulation are not clear. Decreased expression of the glucocorticoid receptor (GR) gene, which is also susceptible to epigenetic modulation, is a strong indicator of impaired HPA axis control. In the context of teenage suicide-completers, we have systematically analyzed the 5'UTR of the GR gene to determine the expression levels of all GR exon-1 transcript variants and their epigenetic state. We also measured the expression and the epigenetic state of the FK506-binding protein 51 (FKBP5/FKBP51), an important modulator of GR activity. Furthermore, steady-state DNA methylation levels depend upon the interplay between enzymes that promote DNA methylation and demethylation activities, thus we analyzed DNA methyltransferases (DNMTs), ten-eleven translocation enzymes (TETs), and growth arrest- and DNA-damage-inducible proteins (GADD45). Focusing on both the prefrontal cortex (PFC) and hippocampus, our results show decreased expression in specific GR exon-1 variants and a strong correlation of DNA methylation changes with gene expression in the PFC. FKBP5 expression is also increased in both areas suggesting a decreased GR sensitivity to cortisol binding. We also identified aberrant expression of DNA methylating and demethylating enzymes in both brain regions. These findings enhance our understanding of the complex transcriptional regulation of GR, providing evidence of epigenetically mediated reprogramming of the GR gene, which could lead to possible epigenetic influences that result in lasting modifications underlying an individual's overall HPA axis response and resilience to stress.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36781843',
'doi' => '10.1038/s41398-023-02345-1',
'modified' => '2023-04-14 09:26:37',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '4675',
'name' => 'Bridging biological cfDNA features and machine learning approaches.',
'authors' => 'Moser T. et al.',
'description' => '<p>Liquid biopsies (LBs), particularly using circulating tumor DNA (ctDNA), are expected to revolutionize precision oncology and blood-based cancer screening. Recent technological improvements, in combination with the ever-growing understanding of cell-free DNA (cfDNA) biology, are enabling the detection of tumor-specific changes with extremely high resolution and new analysis concepts beyond genetic alterations, including methylomics, fragmentomics, and nucleosomics. The interrogation of a large number of markers and the high complexity of data render traditional correlation methods insufficient. In this regard, machine learning (ML) algorithms are increasingly being used to decipher disease- and tissue-specific signals from cfDNA. Here, we review recent insights into biological ctDNA features and how these are incorporated into sophisticated ML applications.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36792446',
'doi' => '10.1016/j.tig.2023.01.004',
'modified' => '2023-04-14 09:28:00',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '4823',
'name' => 'Gene body DNA hydroxymethylation restricts the magnitude oftranscriptional changes during aging.',
'authors' => 'Occean J. R. et al.',
'description' => '<p>DNA hydroxymethylation (5hmC) is the most abundant oxidative derivative of DNA methylation (5mC) and is typically enriched at enhancers and gene bodies of transcriptionally active and tissue-specific genes. Although aberrant genomic 5hmC has been implicated in many age-related diseases, the functional role of the modification in aging remains largely unknown. Here, we report that 5hmC is stably enriched in multiple aged organs. Using the liver and cerebellum as model organs, we show that 5hmC accumulates in gene bodies associated with tissue-specific function and thereby restricts the magnitude of gene expression changes during aging. Mechanistically, we found that 5hmC decreases binding affinity of splicing factors compared to unmodified cytosine and 5mC, and is correlated with age-related alternative splicing events, suggesting RNA splicing as a potential mediator of 5hmC’s transcriptionally restrictive function. Furthermore, we show that various age-related contexts, such as prolonged quiescence and senescence, are partially responsible for driving the accumulation of 5hmC with age. We provide evidence that this age-related function is conserved in mouse and human tissues, and further show that the modification is altered by regimens known to modulate lifespan. Our findings reveal that 5hmC is a regulator of tissue-specific function and may play a role in regulating longevity.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36824863',
'doi' => '10.1101/2023.02.15.528714',
'modified' => '2023-06-14 08:39:26',
'created' => '2023-06-13 22:16:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '4631',
'name' => 'Consistent DNA Hypomethylations in Prostate Cancer.',
'authors' => 'Araúzo-Bravo M.J. et al.',
'description' => '<p>With approximately 1.4 million men annually diagnosed with prostate cancer (PCa) worldwide, PCa remains a dreaded threat to life and source of devastating morbidity. In recent decades, a significant decrease in age-specific PCa mortality has been achieved by increasing prostate-specific antigen (PSA) screening and improving treatments. Nevertheless, upcoming, augmented recommendations against PSA screening underline an escalating disproportion between the benefit and harm of current diagnosis/prognosis and application of radical treatment standards. Undoubtedly, new potent diagnostic and prognostic tools are urgently needed to alleviate this tensed situation. They should allow a more reliable early assessment of the upcoming threat, in order to enable applying timely adjusted and personalized therapy and monitoring. Here, we present a basic study on an epigenetic screening approach by Methylated DNA Immunoprecipitation (MeDIP). We identified genes associated with hypomethylated CpG islands in three PCa sample cohorts. By adjusting our computational biology analyses to focus on single CpG-enriched 60-nucleotide-long DNA probes, we revealed numerous consistently differential methylated DNA segments in PCa. They were associated among other genes with and . These can be used for early discrimination, and might contribute to a new epigenetic tumor classification system of PCa. Our analysis shows that we can dissect short, differential methylated CpG-rich DNA fragments and combinations of them that are consistently present in all tumors. We name them tumor cell-specific differential methylated CpG dinucleotide signatures (TUMS).</p>',
'date' => '2022-12-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36613831',
'doi' => '10.3390/ijms24010386',
'modified' => '2023-03-28 09:03:47',
'created' => '2023-02-21 09:59:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '4534',
'name' => 'RNA 5-Methylcytosine Modification Regulates VegetativeDevelopment Associated with H3K27 Trimethylation inArabidopsis.',
'authors' => 'Zhang D.et al.',
'description' => '<p>Methylating RNA post-transcriptionally is emerging as a significant mechanism of gene regulation in eukaryotes. The crosstalk between RNA methylation and histone modification is critical for chromatin state and gene expression in mammals. However, it is not well understood mechanistically in plants. Here, the authors report a genome-wide correlation between RNA 5-cytosine methylation (m C) and histone 3 lysine27 trimethylation (H3K27me3) in Arabidopsis. The plant-specific Polycomb group (PcG) protein EMBRYONIC FLOWER1 (EMF1) plays dual roles as activators or repressors. Transcriptome-wide RNA m C profiling revealed that m C peaks are mostly enriched in chromatin regions that lacked H3K27me3 in both wild type and emf1 mutants. EMF1 repressed the expression of m C methyltransferase tRNA specific methyltransferase 4B (TRM4B) through H3K4me3, independent of PcG-mediated H3K27me3 mechanism. The 5-Cytosine methylation on targets is increased in emf1 mutants, thereby decreased the mRNA transcripts of photosynthesis and chloroplast genes. In addition, impairing EMF1 activity reduced H3K27me3 levels of PcG targets, such as starch genes, which are de-repressed in emf1 mutants. Both EMF1-mediated promotion and repression of gene activities via m C and H3K27me3 are required for normal vegetative growth. Collectively, t study reveals a previously undescribed epigenetic mechanism of RNA m C modifications and histone modifications to regulate gene expression in eukaryotes.</p>',
'date' => '2022-11-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36382558',
'doi' => '10.1002/advs.202204885',
'modified' => '2022-11-24 08:57:31',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '4541',
'name' => 'Cardiac epigenetic changes in VEGF signaling genes associates with myocardial microvascular rarefaction in experimental chronic kidney disease.',
'authors' => 'Eirin Alfonso and Chade Alejandro R',
'description' => '<p>BACKGROUND: Chronic kidney disease (CKD) is common in patients with heart failure, and often results in left ventricular diastolic dysfunction (LVDD). However, the mechanisms responsible for cardiac damage in CKD-LVDD remain to be elucidated. Epigenetic alterations may impose long-lasting effects on cellular transcription and function, but their exact role in CKD-LVDD is unknown. We investigate whether changes in cardiac site-specific DNA methylation profiles might be implicated in cardiac abnormalities in CKD-LVDD. METHODS: CKD-LVDD and normal control pigs (n=6 each) were studied for 14 weeks. Renal and cardiac hemodynamics were quantified by multidetector CT and echocardiography. In randomly selected pigs (n=3/group), cardiac site-specific 5-methylcytosine (5mC) immunoprecipitation (MeDIP)- and mRNA-sequencing (seq) was performed, followed by integrated (MeDiP-seq/mRNA-seq analysis), and confirmatory ex vivo studies. RESULTS: MeDIP-seq analysis revealed 261 genes with higher (fold-change>1.4; p<0.05) and 162 genes with lower (fold-change<0.7; p<0.05) 5mC levels in CKD-LVDD versus normal pigs, which were primarily implicated in vascular endothelial growth factor (VEGF)-related signaling and angiogenesis. Integrated MeDiP-seq/mRNA-seq analysis identified a select group of VEGF-related genes in which 5mC levels were higher, but mRNA expression lower in CKD-LVDD versus normal pigs. Cardiac VEGF signaling gene and VEGF protein expression was blunted in CKD-LVDD compared to controls and associated with decreased subendocardial microvascular density. CONCLUSIONS: Cardiac epigenetic changes in VEGF-related genes are associated with impaired angiogenesis and cardiac microvascular rarefaction in swine CKD-LVDD. These observations may assist in developing novel therapies to ameliorate cardiac damage in CKD-LVDD.</p>',
'date' => '2022-11-01',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/36367693/',
'doi' => '10.1152/ajpheart.00522.2022',
'modified' => '2022-11-25 09:03:31',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '4511',
'name' => 'The Arabidopsis APOLO and human UPAT sequence-unrelated longnoncoding RNAs can modulate DNA and histone methylation machineries inplants.',
'authors' => 'Fonouni-Farde C. et al.',
'description' => '<p>BACKGROUND: RNA-DNA hybrid (R-loop)-associated long noncoding RNAs (lncRNAs), including the Arabidopsis lncRNA AUXIN-REGULATED PROMOTER LOOP (APOLO), are emerging as important regulators of three-dimensional chromatin conformation and gene transcriptional activity. RESULTS: Here, we show that in addition to the PRC1-component LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), APOLO interacts with the methylcytosine-binding protein VARIANT IN METHYLATION 1 (VIM1), a conserved homolog of the mammalian DNA methylation regulator UBIQUITIN-LIKE CONTAINING PHD AND RING FINGER DOMAINS 1 (UHRF1). The APOLO-VIM1-LHP1 complex directly regulates the transcription of the auxin biosynthesis gene YUCCA2 by dynamically determining DNA methylation and H3K27me3 deposition over its promoter during the plant thermomorphogenic response. Strikingly, we demonstrate that the lncRNA UHRF1 Protein Associated Transcript (UPAT), a direct interactor of UHRF1 in humans, can be recognized by VIM1 and LHP1 in plant cells, despite the lack of sequence homology between UPAT and APOLO. In addition, we show that increased levels of APOLO or UPAT hamper VIM1 and LHP1 binding to YUCCA2 promoter and globally alter the Arabidopsis transcriptome in a similar manner. CONCLUSIONS: Collectively, our results uncover a new mechanism in which a plant lncRNA coordinates Polycomb action and DNA methylation through the interaction with VIM1, and indicates that evolutionary unrelated lncRNAs with potentially conserved structures may exert similar functions by interacting with homolog partners.</p>',
'date' => '2022-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36038910',
'doi' => '10.1186/s13059-022-02750-7',
'modified' => '2022-11-21 10:43:16',
'created' => '2022-11-15 09:26:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '4438',
'name' => 'A genome-wide screen reveals new regulators of the 2-cell-like cell state',
'authors' => 'Defossez Pierre-Antoine et al.',
'description' => '<p>In mammals, only the zygote and blastomeres of the early embryo are fully totipotent. This totipotency is mirrored in vitro by mouse "2-cell-like cells" (2CLCs), which appear at low frequency in cultures of Embryonic Stem cells (ESCs). Because totipotency is incompletely understood, we carried out a genomewide CRISPR KO screen in mouse ESCs, searching for mutants that reactivate the expression of Dazl, a robust 2-cell-like marker. Using secondary screens, we identify four mutants that reactivate not just Dazl, but also a broader 2-cell-like signature: the E3 ubiquitin ligase adaptor SPOP, the Zinc Finger transcription factor ZBTB14, MCM3AP, a component of the RNA processing complex TREX-2, and the lysine demethylase KDM5C. Functional experiments show how these factors link to known players of the 2 celllike state. These results extend our knowledge of totipotency, a key phase of organismal life.</p>',
'date' => '2022-06-01',
'pmid' => 'https://doi.org/10.21203%2Frs.3.rs-1561018%2Fv1',
'doi' => '10.21203/rs.3.rs-1561018/v1',
'modified' => '2022-09-28 09:23:42',
'created' => '2022-09-08 16:32:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '4553',
'name' => 'NSUN2-mediated RNA mC modification modulates uveal melanoma cellproliferation and migration.',
'authors' => 'Luo Guangying et al.',
'description' => '<p>RNA 5-methylcytosine (mC) is a widespread post-transcriptional modification involved in diverse biological processes through controlling RNA metabolism. However, its roles in uveal melanoma (UM) remain unknown. Here, we describe the biological roles and regulatory mechanisms of RNA mC in UM. Initially, we identified significantly elevated global RNA mC levels in both UM cells and tissue specimens using ELISA assay and dot blot analysis. Meanwhile, NOP2/Sun RNA methyltransferase family member 2 (NSUN2) was upregulated in both types of these samples, whereas NSUN2 knockdown significantly decreased RNA mC level. Such declines inhibited UM cell migration and suppressed cell proliferation through cell cycle G1 arrest. Furthermore, bioinformatic analyses, mC-RIP-qPCR, and luciferase assay identified β-Catenin (CTNNB1) as a direct target of NSUN2-mediated mC modification in UM cells. Additionally, overexpression of miR-124a in UM cells diminished NSUN2 expression levels indicating that it is an upstream regulator of this response. Our study suggests that NSUN2-mediated RNA mC methylation provides a potential novel target to improve the therapeutic management of UM pathogenesis.</p>',
'date' => '2022-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/35757999',
'doi' => '10.1080/15592294.2022.2088047',
'modified' => '2022-11-24 10:14:24',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '4340',
'name' => 'Global DNA methylation and cellular 5-methylcytosine and H4acetylated patterns in primary and secondary dormant seeds of Capsellabursa-pastoris (L.) Medik. (shepherd's purse).',
'authors' => 'Gomez-Cabellos Sara et al.',
'description' => '<p>Despite the importance of dormancy and dormancy cycling for plants' fitness and life cycle phenology, a comprehensive characterization of the global and cellular epigenetic patterns across space and time in different seed dormancy states is lacking. Using Capsella bursa-pastoris (L.) Medik. (shepherd's purse) seeds with primary and secondary dormancy, we investigated the dynamics of global genomic DNA methylation and explored the spatio-temporal distribution of 5-methylcytosine (5-mC) and histone H4 acetylated (H4Ac) epigenetic marks. Seeds were imbibed at 30 °C in a light regime to maintain primary dormancy, or in darkness to induce secondary dormancy. An ELISA-based method was used to quantify DNA methylation, in relation to total genomic cytosines. Immunolocalization of 5-mC and H4Ac within whole seeds (i.e., including testa) was assessed with reference to embryo anatomy. Global DNA methylation levels were highest in prolonged (14 days) imbibed primary dormant seeds, with more 5-mC marked nuclei present only in specific parts of the seed (e.g., SAM and cotyledons). In secondary dormant seeds, global methylation levels and 5-mC signal where higher at 3 and 7 days than 1 or 14 days. With respect to acetylation, seeds had fewer H4Ac marked nuclei (e.g., SAM) in deeper dormant states, for both types of dormancy. However, the RAM still showed signal after 14 days of imbibition under dormancy-inducing conditions, suggesting a central role for the radicle/RAM in the response to perceived ambient changes and the adjustment of the seed dormancy state. Thus, we show that seed dormancy involves extensive cellular remodeling of DNA methylation and H4 acetylation.</p>',
'date' => '2022-05-01',
'pmid' => 'https://doi.org/10.1007%2Fs00709-021-01678-2',
'doi' => '10.1007/s00709-021-01678-2',
'modified' => '2022-06-20 09:19:49',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '4404',
'name' => 'Stella regulates the Development of Female Germline Stem Cells byModulating Chromatin Structure and DNA Methylation.',
'authors' => 'Hou Changliang et al.',
'description' => '<p>Female germline stem cells (FGSCs) have the ability to self-renew and differentiate into oocytes. , encoded by a maternal effect gene, plays an important role in oogenesis and early embryonic development. However, its function in FGSCs remains unclear. In this study, we showed that CRISPR/Cas9-mediated knockout of promoted FGSC proliferation and reduced the level of genome-wide DNA methylation of FGSCs. Conversely, overexpression led to the opposite results, and enhanced FGSC differentiation. We also performed an integrative analysis of chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), high-throughput genome-wide chromosome conformation capture (Hi-C), and use of our published epigenetic data. Results indicated that the binding sites of STELLA and active histones H3K4me3 and H3K27ac were enriched near the TAD boundaries. Hi-C analysis showed that overexpression attenuated the interaction within TADs, and interestingly enhanced the TAD boundary strength in STELLA-associated regions. Taking these findings together, our study not only reveals the role of in regulating DNA methylation and chromatin structure, but also provides a better understanding of FGSC development.</p>',
'date' => '2022-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9066111/',
'doi' => '10.7150/ijbs.69240',
'modified' => '2022-08-11 14:54:29',
'created' => '2022-08-11 12:14:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '4327',
'name' => 'Highly recurrent epimutations in gastric cancer CpG islandmethylator phenotypes and inflammation',
'authors' => 'Padmanabhan N. et al.',
'description' => '<p>Background CIMP (CpG island methylator phenotype) is an epigenetic molecular subtype, observed in multiple malignancies and associated with the epigenetic silencing of tumor suppressors. Currently, for most cancers including gastric cancer (GC), mechanisms underlying CIMP remain poorly understood. We sought to discover molecular contributors to CIMP in GC, by performing global DNA methylation, gene expression, and proteomics profiling across 14 gastric cell lines, followed by similar integrative analysis in 50 GC cell lines and 467 primary GCs. Results We identify the cystathionine beta-synthase enzyme (CBS) as a highly recurrent target of epigenetic silencing in CIMP GC. Likewise, we show that CBS epimutations are significantly associated with CIMP in various other cancers, occurring even in premalignant gastroesophageal conditions and longitudinally linked to clinical persistence. Of note, CRISPR deletion of CBS in normal gastric epithelial cells induces widespread DNA methylation changes that overlap with primary GC CIMP patterns. Reflecting its metabolic role as a gatekeeper interlinking the methionine and homocysteine cycles, CBS loss in vitro also causes reductions in the anti-inflammatory gasotransmitter hydrogen sulfide (H2S), with concomitant increase in NF-κB activity. In a murine genetic model of CBS deficiency, preliminary data indicate upregulated immune-mediated transcriptional signatures in the stomach. Conclusions Our results implicate CBS as a bi-faceted modifier of aberrant DNA methylation and inflammation in GC and highlights H2S donors as a potential new therapy for CBS-silenced lesions. Supplementary Information The online version contains supplementary material available at 10.1186/s13059-021-02375-2.</p>',
'date' => '2021-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/34074348',
'doi' => '10.1186/s13059-021-02375-2',
'modified' => '2022-08-03 16:01:40',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '4336',
'name' => 'LINE-1 transcription in round spermatids is associated with accretion of5-carboxylcytosine in their open reading frames',
'authors' => 'Blythe M. et al. ',
'description' => '<p>Chromatin of male and female gametes undergoes a number of reprogramming events during the transition from germ cell to embryonic developmental programs. Although the rearrangement of DNA methylation patterns occurring in the zygote has been extensively characterized, little is known about the dynamics of DNA modifications during spermatid maturation. Here, we demonstrate that the dynamics of 5-carboxylcytosine (5caC) correlate with active transcription of LINE-1 retroelements during murine spermiogenesis. We show that the open reading frames of active and evolutionary young LINE-1s are 5caC-enriched in round spermatids and 5caC is eliminated from LINE-1s and spermiogenesis-specific genes during spermatid maturation, being simultaneously retained at promoters and introns of developmental genes. Our results reveal an association of 5caC with activity of LINE-1 retrotransposons suggesting a potential direct role for this DNA modification in fine regulation of their transcription.</p>',
'date' => '2021-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/34099857',
'doi' => '10.1038/s42003-021-02217-8',
'modified' => '2022-08-03 16:17:04',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '4150',
'name' => 'Sensitive and reproducible cell-free methylome quantification with synthetic spike-in controls',
'authors' => 'Wilson, S.L. et al.',
'description' => '<p>Background. Cell-free methylated DNA immunoprecipitation-sequencing (cfMeDIP-seq) identifies genomic regions with DNA methylation, using a protocol adapted to work with low-input DNA samples and with cell-free DNA (cfDNA). This method allows for DNA methylation profiling of circulating tumour DNA in cancer patients’ blood samples. Such epigenetic profiling of circulating tumour DNA provides information about in which tissues tumour DNA originates, a key requirement of any test for early cancer detection. In addition, DNA methylation signatures provide prognostic information and can detect relapse. For robust quantitative comparisons between samples, immunoprecipitation enrichment methods like cfMeDIP-seq require normalization against common reference controls.</p>',
'date' => '2021-04-01',
'pmid' => 'https://doi.org/10.1101%2F2021.02.12.430289',
'doi' => '10.1101/2021.02.12.430289',
'modified' => '2022-01-13 15:16:23',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '3985',
'name' => 'Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes.',
'authors' => 'Nuzzo PV, Berchuck JE, Korthauer K, Spisak S, Nassar AH, Abou Alaiwi S, Chakravarthy A, Shen SY, Bakouny Z, Boccardo F, Steinharter J, Bouchard G, Curran CR, Pan W, Baca SC, Seo JH, Lee GM, Michaelson MD, Chang SL, Waikar SS, Sonpavde G, Irizarry RA, Pome',
'description' => '<p>Improving early cancer detection has the potential to substantially reduce cancer-related mortality. Cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) is a highly sensitive assay capable of detecting early-stage tumors. We report accurate classification of patients across all stages of renal cell carcinoma (RCC) in plasma (area under the receiver operating characteristic (AUROC) curve of 0.99) and demonstrate the validity of this assay to identify patients with RCC using urine cell-free DNA (cfDNA; AUROC of 0.86).</p>',
'date' => '2020-06-22',
'pmid' => 'http://www.pubmed.gov/32572266',
'doi' => '10.1038/s41591-020-0933-1',
'modified' => '2020-09-01 15:13:49',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '3984',
'name' => 'Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes.',
'authors' => 'Nassiri F, Chakravarthy A, Feng S, Shen SY, Nejad R, Zuccato JA, Voisin MR, Patil V, Horbinski C, Aldape K, Zadeh G, De Carvalho DD',
'description' => '<p>Definitive diagnosis of intracranial tumors relies on tissue specimens obtained by invasive surgery. Noninvasive diagnostic approaches provide an opportunity to avoid surgery and mitigate unnecessary risk to patients. In the present study, we show that DNA-methylation profiles from plasma reveal highly specific signatures to detect and accurately discriminate common primary intracranial tumors that share cell-of-origin lineages and can be challenging to distinguish using standard-of-care imaging.</p>',
'date' => '2020-06-22',
'pmid' => 'http://www.pubmed.gov/32572265',
'doi' => '10.1038/s41591-020-0932-2',
'modified' => '2020-09-01 15:14:45',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '4030',
'name' => 'AXR1 affects DNA methylation independently of its role in regulatingmeiotic crossover localization.',
'authors' => 'Christophorou, N and She, W and Long, J and Hurel, A and Beaubiat, S andIdir, Y and Tagliaro-Jahns, M and Chambon, A and Solier, V and Vezon, D andGrelon, M and Feng, X and Bouché, N and Mézard, C',
'description' => '<p>Meiotic crossovers (COs) are important for reshuffling genetic information between homologous chromosomes and they are essential for their correct segregation. COs are unevenly distributed along chromosomes and the underlying mechanisms controlling CO localization are not well understood. We previously showed that meiotic COs are mis-localized in the absence of AXR1, an enzyme involved in the neddylation/rubylation protein modification pathway in Arabidopsis thaliana. Here, we report that in axr1-/-, male meiocytes show a strong defect in chromosome pairing whereas the formation of the telomere bouquet is not affected. COs are also redistributed towards subtelomeric chromosomal ends where they frequently form clusters, in contrast to large central regions depleted in recombination. The CO suppressed regions correlate with DNA hypermethylation of transposable elements (TEs) in the CHH context in axr1-/- meiocytes. Through examining somatic methylomes, we found axr1-/- affects DNA methylation in a plant, causing hypermethylation in all sequence contexts (CG, CHG and CHH) in TEs. Impairment of the main pathways involved in DNA methylation is epistatic over axr1-/- for DNA methylation in somatic cells but does not restore regular chromosome segregation during meiosis. Collectively, our findings reveal that the neddylation pathway not only regulates hormonal perception and CO distribution but is also, directly or indirectly, a major limiting pathway of TE DNA methylation in somatic cells.</p>',
'date' => '2020-06-01',
'pmid' => 'http://www.pubmed.gov/32598340',
'doi' => '10.1371/journal.',
'modified' => '2020-12-16 17:58:51',
'created' => '2020-10-12 14:54:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '3951',
'name' => 'In vitro capture and characterization of embryonic rosette-stage pluripotency between naive and primed states.',
'authors' => 'Neagu A, van Genderen E, Escudero I, Verwegen L, Kurek D, Lehmann J, Stel J, Dirks RAM, van Mierlo G, Maas A, Eleveld C, Ge Y, den Dekker AT, Brouwer RWW, van IJcken WFJ, Modic M, Drukker M, Jansen JH, Rivron NC, Baart EB, Marks H, Ten Berge D',
'description' => '<p>Following implantation, the naive pluripotent epiblast of the mouse blastocyst generates a rosette, undergoes lumenogenesis and forms the primed pluripotent egg cylinder, which is able to generate the embryonic tissues. How pluripotency progression and morphogenesis are linked and whether intermediate pluripotent states exist remain controversial. We identify here a rosette pluripotent state defined by the co-expression of naive factors with the transcription factor OTX2. Downregulation of blastocyst WNT signals drives the transition into rosette pluripotency by inducing OTX2. The rosette then activates MEK signals that induce lumenogenesis and drive progression to primed pluripotency. Consequently, combined WNT and MEK inhibition supports rosette-like stem cells, a self-renewing naive-primed intermediate. Rosette-like stem cells erase constitutive heterochromatin marks and display a primed chromatin landscape, with bivalently marked primed pluripotency genes. Nonetheless, WNT induces reversion to naive pluripotency. The rosette is therefore a reversible pluripotent intermediate whereby control over both pluripotency progression and morphogenesis pivots from WNT to MEK signals.</p>',
'date' => '2020-05-01',
'pmid' => 'http://www.pubmed.gov/32367046',
'doi' => '10.1038/s41556-020-0508-x',
'modified' => '2020-08-17 09:55:37',
'created' => '2020-08-10 12:12:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '3834',
'name' => 'Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.',
'authors' => 'Liu J, An Z, Luo J, Li J, Li F, Zhang Z',
'description' => '<p>MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of transcriptome-wide profile and potential function of m5C in splicing remains to be elucidated due to lack of isoform level m5C quantification tool. RESULTS: We developed a computational package to quantify Epitranscriptomal RNA m5C at the transcript isoform level (named Episo). Episo consists of three tools, mapper, quant and Bisulfitefq, for mapping, quantifying, and simulating RNA-BisSeq data, respectively. The high accuracy of Episo was validated using an improved m5C-specific methylated RNA immunoprecipitation (meRIP) protocol, as well as a set of in silico experiments. By applying Episo to public human and mouse RNA-BisSeq data, we found that the RNA m5C is not evenly distributed among the transcript isoforms, implying the m5C may subject to be regulated at isoform level. AVAILABILITY: Episo is released under the GNU GPLv3+ license. The resource code Episo is freely accessible from https://github.com/liujunfengtop/Episo (with Tophat/cufflink) and https://github.com/liujunfengtop/Episo/tree/master/Episo_Kallisto (with Kallisto). SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.</p>',
'date' => '2019-12-03',
'pmid' => 'http://www.pubmed.gov/31794005',
'doi' => '10.1093/bioinformatics/btz900/5651015',
'modified' => '2020-02-25 13:26:22',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 25 => array(
'id' => '3827',
'name' => 'Intra- and inter-generational changes in the cortical DNA methylome in response to therapeutic intermittent hypoxia in mice.',
'authors' => 'Belmonte KCD, Harman JC, Lanson NA, Gidday JM',
'description' => '<p>Recent evidence from our lab documents functional resilience to retinal ischemic injury in untreated mice derived from parents exposed to repetitive hypoxic conditioning (RHC) prior to breeding. To begin to understand the epigenetic basis of this intergenerational protection, we used methylated DNA immunoprecipitation and sequencing to identify genes with differentially-methylated promoters (DMGPs) in the prefrontal cortex of mice treated directly with the same RHC stimulus (F0-RHC), and in the prefrontal cortex of their untreated F1-generation offspring (F1-*RHC). Subsequent bioinformatic analyses provided key mechanistic insights into how changes in gene expression secondary to promoter hypo- and hyper-methylation might afford such protection within and across generations. We found extensive changes in DNA methylation in both generations consistent with the expression of many survival-promoting genes, with twice the number of DMGPs in the cortex of F1*RHC mice relative to their F0 parents that were directly exposed to RHC. In contrast to our hypothesis that similar epigenetic modifications would be realized in the cortices of both F0-RHC and F1-*RHC mice, we instead found relatively few DMGPs common to both generations; in fact, each generation manifested expected injury resilience via distinctly unique gene expression profiles. Whereas in the cortex of F0-RHC mice, predicted protein-protein interactions reflected the activation of an anti-ischemic phenotype, networks activated in F1-*RHC cortex comprised networks indicative of a much broader cytoprotective phenotype. Altogether, our results suggest that the intergenerational transfer of an acquired phenotype to offspring does not necessarily require the faithful recapitulation of the conditioning-modified DNA methylome of the parent.</p>',
'date' => '2019-11-25',
'pmid' => 'http://www.pubmed.gov/31762411',
'doi' => '10.1152/physiolgenomics.00094.2019',
'modified' => '2020-02-25 13:35:09',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 26 => array(
'id' => '3814',
'name' => 'Lithium treatment reverses irradiation-induced changes in rodent neural progenitors and rescues cognition.',
'authors' => 'Zanni G, Goto S, Fragopoulou AF, Gaudenzi G, Naidoo V, Di Martino E, Levy G, Dominguez CA, Dethlefsen O, Cedazo-Minguez A, Merino-Serrais P, Stamatakis A, Hermanson O, Blomgren K',
'description' => '<p>Cranial radiotherapy in children has detrimental effects on cognition, mood, and social competence in young cancer survivors. Treatments harnessing hippocampal neurogenesis are currently of great relevance in this context. Lithium, a well-known mood stabilizer, has both neuroprotective, pro-neurogenic as well as antitumor effects, and in the current study we introduced lithium treatment 4 weeks after irradiation. Female mice received a single 4 Gy whole-brain radiation dose on postnatal day (PND) 21 and were randomized to 0.24% Li2CO chow or normal chow from PND 49 to 77. Hippocampal neurogenesis was assessed on PND 77, 91, and 105. We found that lithium treatment had a pro-proliferative effect on neural progenitors, but neuronal integration occurred only after it was discontinued. Also, the treatment ameliorated deficits in spatial learning and memory retention observed in irradiated mice. Gene expression profiling and DNA methylation analysis identified two novel factors related to the observed effects, Tppp, associated with microtubule stabilization, and GAD2/65, associated with neuronal signaling. Our results show that lithium treatment reverses irradiation-induced loss of hippocampal neurogenesis and cognitive impairment even when introduced long after the injury. We propose that lithium treatment should be intermittent in order to first make neural progenitors proliferate and then, upon discontinuation, allow them to differentiate. Our findings suggest that pharmacological treatment of cognitive so-called late effects in childhood cancer survivors is possible.</p>',
'date' => '2019-11-14',
'pmid' => 'http://www.pubmed.gov/31723242',
'doi' => '10.1038/s41380-019-0584-0',
'modified' => '2019-12-05 10:58:44',
'created' => '2019-12-02 15:25:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 27 => array(
'id' => '3773',
'name' => 'Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA.',
'authors' => 'Shen SY, Burgener JM, Bratman SV, De Carvalho DD',
'description' => '<p>Circulating cell-free DNA (cfDNA) comprises small DNA fragments derived from normal and tumor tissue that are released into the bloodstream. Recently, methylation profiling of cfDNA as a liquid biopsy tool has been gaining prominence due to the presence of tissue-specific markers in cfDNA. We have previously reported cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) as a sensitive, low-input, cost-efficient and bisulfite-free approach to profiling DNA methylomes of plasma cfDNA. cfMeDIP-seq is an extension of a previously published MeDIP-seq protocol and is adapted to allow for methylome profiling of samples with low input (ranging from 1 to 10 ng) of DNA, which is enabled by the addition of 'filler DNA' before immunoprecipitation. This protocol is not limited to plasma cfDNA; it can also be applied to other samples that are naturally sheared and at low availability (e.g., urinary cfDNA and cerebrospinal fluid cfDNA), and is potentially applicable to other applications beyond cancer detection, including prenatal diagnostics, cardiology and monitoring of immune response. The protocol presented here should enable any standard molecular laboratory to generate cfMeDIP-seq libraries from plasma cfDNA in ~3-4 d.</p>',
'date' => '2019-08-30',
'pmid' => 'http://www.pubmed.gov/31471598',
'doi' => '10.1038/s41596-019-0202-2',
'modified' => '2019-10-02 17:07:45',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 28 => array(
'id' => '3763',
'name' => 'Silencing of tumor-suppressive NR_023387 in renal cell carcinoma via promoter hypermethylation and HNF4A deficiency.',
'authors' => 'Zhou H, Guo L, Yao W, Shi R, Yu G, Xu H, Ye Z',
'description' => '<p>Dysregulation of the epigenetic status of long noncoding RNAs (lncRNAs) has been linked to diverse human diseases including human cancers. However, the landscape of the whole-genome methylation profile of lncRNAs and the precise roles of these lncRNAs remain elusive in renal cell carcinoma (RCC). We first examined lncRNA expression profiles in RCC tissues and corresponding adjacent normal tissues (NTs) to identify the lncRNA signature of RCC, then lncRNA Promoter Microarray was performed to depict the whole-genome methylation profile of lncRNAs in RCC. Combined analysis of the lncRNAs expression profiles and lncRNAs Promoter Microarray identified a series of downregulated lncRNAs with hypermethylated promoter regions, including NR_023387. Quantitative real-time polymerase chain reaction (RT-PCR) implied that NR_023387 was significantly downregulated in RCC tissues and cell lines, and lower expression of NR_023387 was correlated with shorter overall survival. Methylation-specific PCR, MassARRAY, and demethylation drug treatment indicated that hypermethylation in the NR_023387 promoter contributed to its silencing in RCC. Besides, HNF4A regulated the expression of NR_023387 via transcriptional activation. Functional experiments demonstrated NR_023387 exerted tumor-suppressive roles in RCC via suppressing the proliferation, migration, invasion, tumor growth, and metastasis of RCC. Furthermore, we identified MGP as a putative downstream molecule of NR_023387, which promoted the epithelial-mesenchymal transition of RCC cells. Our study provides the first whole-genome lncRNA methylation profile in RCC. Our combined analysis identifies a tumor-suppressive and prognosis-related lncRNA NR_023387, which is silenced in RCC via promoter hypermethylation and HNF4A deficiency, and may exert its tumor-suppressive roles by downregulating the oncogenic MGP.</p>',
'date' => '2019-08-20',
'pmid' => 'http://www.pubmed.gov/31432508',
'doi' => '10.1002/jcp.29115',
'modified' => '2019-10-03 10:02:27',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 29 => array(
'id' => '3770',
'name' => 'Epitranscriptomic Addition of mC to HIV-1 Transcripts Regulates Viral Gene Expression.',
'authors' => 'Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR',
'description' => '<p>How the covalent modification of mRNA ribonucleotides, termed epitranscriptomic modifications, alters mRNA function remains unclear. One issue has been the difficulty of quantifying these modifications. Using purified HIV-1 genomic RNA, we show that this RNA bears more epitranscriptomic modifications than the average cellular mRNA, with 5-methylcytosine (mC) and 2'O-methyl modifications being particularly prevalent. The methyltransferase NSUN2 serves as the primary writer for mC on HIV-1 RNAs. NSUN2 inactivation inhibits not only mC addition to HIV-1 transcripts but also viral replication. This inhibition results from reduced HIV-1 protein, but not mRNA, expression, which in turn correlates with reduced ribosome binding to viral mRNAs. In addition, loss of mC dysregulates the alternative splicing of viral RNAs. These data identify mC as a post-transcriptional regulator of both splicing and function of HIV-1 mRNA, thereby affecting directly viral gene expression.</p>',
'date' => '2019-08-14',
'pmid' => 'http://www.pubmed.gov/31415754',
'doi' => '10.1016/j.chom.2019.07.005',
'modified' => '2019-10-03 09:18:50',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 30 => array(
'id' => '3741',
'name' => 'Aberrant expression of imprinted lncRNA MEG8 causes trophoblast dysfunction and abortion.',
'authors' => 'Sheng F, Sun N, Ji Y, Ma Y, Ding H, Zhang Q, Yang F, Li W',
'description' => '<p>Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs whose nucleotides are longer than 200 bp. Previous studies have shown that they play an important regulatory role in many developmental processes and biological pathways. However, the contributions of lncRNAs to placental development are largely unknown. Here, our study aimed to investigate the lncRNA expression signatures in placental development by performing a microarray lncRNA screen. Placental samples were obtained from pregnant C57BL/6 female mice at three key developmental time points (embryonic day E7.5, E13.5, and E19.5). Microarrays were used to analyze the differential expression of lncRNAs during placental development. In addition to the genomic imprinting region and the dynamic DNA methylation status during placental development, we screened imprinted lncRNAs whose expression was controlled by DNA methylation during placental development. We found that the imprinted lncRNA Rian may play an important role during placental development. Its homologous sequence lncRNA MEG8 (RIAN) was abnormally highly expressed in human spontaneous abortion villi. Upregulation of MEG8 expression in trophoblast cell lines decreased cell proliferation and invasion, whereas downregulation of MEG8 expression had the opposite effect. Furthermore, DNA methylation results showed that the methylation of the MEG8 promoter region was increased in spontaneous abortion villi. There was dynamic spatiotemporal expression of imprinted lncRNAs during placental development. The imprinted lncRNA MEG8 is involved in the regulation of early trophoblast cell function. Promoter methylation abnormalities can cause trophoblastic cell defects, which may be one of the factors that occurs in early unexplained spontaneous abortion.</p>',
'date' => '2019-07-02',
'pmid' => 'http://www.pubmed.gov/31265183',
'doi' => '10.1002/jcb.29002',
'modified' => '2019-08-06 16:45:53',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 31 => array(
'id' => '3731',
'name' => 'Defining UHRF1 Domains that Support Maintenance of Human Colon Cancer DNA Methylation and Oncogenic Properties.',
'authors' => 'Kong X, Chen J, Xie W, Brown SM, Cai Y, Wu K, Fan D, Nie Y, Yegnasubramanian S, Tiedemann RL, Tao Y, Chiu Yen RW, Topper MJ, Zahnow CA, Easwaran H, Rothbart SB, Xia L, Baylin SB',
'description' => '<p>UHRF1 facilitates the establishment and maintenance of DNA methylation patterns in mammalian cells. The establishment domains are defined, including E3 ligase function, but the maintenance domains are poorly characterized. Here, we demonstrate that UHRF1 histone- and hemimethylated DNA binding functions, but not E3 ligase activity, maintain cancer-specific DNA methylation in human colorectal cancer (CRC) cells. Disrupting either chromatin reader activity reverses DNA hypermethylation, reactivates epigenetically silenced tumor suppressor genes (TSGs), and reduces CRC oncogenic properties. Moreover, an inverse correlation between high UHRF1 and low TSG expression tracks with CRC progression and reduced patient survival. Defining critical UHRF1 domain functions and its relationship with CRC prognosis suggests directions for, and value of, targeting this protein to develop therapeutic DNA demethylating agents.</p>',
'date' => '2019-04-15',
'pmid' => 'http://www.pubmed.gov/30956060',
'doi' => '10.1016/j.ccell.2019.03.003',
'modified' => '2019-08-07 09:14:54',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 32 => array(
'id' => '3729',
'name' => 'Tricarboxylic Acid Cycle Activity and Remodeling of Glycerophosphocholine Lipids Support Cytokine Induction in Response to Fungal Patterns.',
'authors' => 'Márquez S, Fernández JJ, Mancebo C, Herrero-Sánchez C, Alonso S, Sandoval TA, Rodríguez Prados M, Cubillos-Ruiz JR, Montero O, Fernández N, Sánchez Crespo M',
'description' => '<p>Increased glycolysis parallels immune cell activation, but the role of pyruvate remains largely unexplored. We found that stimulation of dendritic cells with the fungal surrogate zymosan causes decreases of pyruvate, citrate, itaconate, and α-ketoglutarate, while increasing oxaloacetate, succinate, lactate, oxygen consumption, and pyruvate dehydrogenase activity. Expression of IL10 and IL23A (the gene encoding the p19 chain of IL-23) depended on pyruvate dehydrogenase activity. Mechanistically, pyruvate reinforced histone H3 acetylation, and acetate rescued the effect of mitochondrial pyruvate carrier inhibition, most likely because it is a substrate of the acetyl-CoA producing enzyme ACSS2. Mice lacking the receptor of the lipid mediator platelet-activating factor (PAF; 1-O-hexadecyl-2-acetyl-sn-glycero-3-phosphocholine) showed reduced production of IL-10 and IL-23 that is explained by the requirement of acetyl-CoA for PAF biosynthesis and its ensuing autocrine function. Acetyl-CoA therefore intertwines fatty acid remodeling of glycerophospholipids and energetic metabolism during cytokine induction.</p>',
'date' => '2019-04-09',
'pmid' => 'http://www.pubmed.gov/30970255',
'doi' => '10.1016/j.celrep.2019.03.033',
'modified' => '2019-08-07 09:15:46',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 33 => array(
'id' => '3693',
'name' => 'Increased Serine and One-Carbon Pathway Metabolism by PKCλ/ι Deficiency Promotes Neuroendocrine Prostate Cancer.',
'authors' => 'Reina-Campos M, Linares JF, Duran A, Cordes T, L'Hermitte A, Badur MG, Bhangoo MS, Thorson PK, Richards A, Rooslid T, Garcia-Olmo DC, Nam-Cha SY, Salinas-Sanchez AS, Eng K, Beltran H, Scott DA, Metallo CM, Moscat J, Diaz-Meco MT',
'description' => '<p>Increasingly effective therapies targeting the androgen receptor have paradoxically promoted the incidence of neuroendocrine prostate cancer (NEPC), the most lethal subtype of castration-resistant prostate cancer (PCa), for which there is no effective therapy. Here we report that protein kinase C (PKC)λ/ι is downregulated in de novo and during therapy-induced NEPC, which results in the upregulation of serine biosynthesis through an mTORC1/ATF4-driven pathway. This metabolic reprogramming supports cell proliferation and increases intracellular S-adenosyl methionine (SAM) levels to feed epigenetic changes that favor the development of NEPC characteristics. Altogether, we have uncovered a metabolic vulnerability triggered by PKCλ/ι deficiency in NEPC, which offers potentially actionable targets to prevent therapy resistance in PCa.</p>',
'date' => '2019-03-18',
'pmid' => 'http://www.pubmed.gov/30827887',
'doi' => '10.1016/j.ccell.2019.01.018',
'modified' => '2019-06-28 13:49:24',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 34 => array(
'id' => '3660',
'name' => 'Global distribution of DNA hydroxymethylation and DNA methylation in chronic lymphocytic leukemia.',
'authors' => 'Wernig-Zorc S, Yadav MP, Kopparapu PK, Bemark M, Kristjansdottir HL, Andersson PO, Kanduri C, Kanduri M',
'description' => '<p>BACKGROUND: Chronic lymphocytic leukemia (CLL) has been a good model system to understand the functional role of 5-methylcytosine (5-mC) in cancer progression. More recently, an oxidized form of 5-mC, 5-hydroxymethylcytosine (5-hmC) has gained lot of attention as a regulatory epigenetic modification with prognostic and diagnostic implications for several cancers. However, there is no global study exploring the role of 5-hydroxymethylcytosine (5-hmC) levels in CLL. Herein, using mass spectrometry and hMeDIP-sequencing, we analysed the dynamics of 5-hmC during B cell maturation and CLL pathogenesis. RESULTS: We show that naïve B-cells had higher levels of 5-hmC and 5-mC compared to non-class switched and class-switched memory B-cells. We found a significant decrease in global 5-mC levels in CLL patients (n = 15) compared to naïve and memory B cells, with no changes detected between the CLL prognostic groups. On the other hand, global 5-hmC levels of CLL patients were similar to memory B cells and reduced compared to naïve B cells. Interestingly, 5-hmC levels were increased at regulatory regions such as gene-body, CpG island shores and shelves and 5-hmC distribution over the gene-body positively correlated with degree of transcriptional activity. Importantly, CLL samples showed aberrant 5-hmC and 5-mC pattern over gene-body compared to well-defined patterns in normal B-cells. Integrated analysis of 5-hmC and RNA-sequencing from CLL datasets identified three novel oncogenic drivers that could have potential roles in CLL development and progression. CONCLUSIONS: Thus, our study suggests that the global loss of 5-hmC, accompanied by its significant increase at the gene regulatory regions, constitute a novel hallmark of CLL pathogenesis. Our combined analysis of 5-mC and 5-hmC sequencing provided insights into the potential role of 5-hmC in modulating gene expression changes during CLL pathogenesis.</p>',
'date' => '2019-01-07',
'pmid' => 'http://www.pubmed.gov/30616658',
'doi' => '10.1186/s13072‑018‑0252‑7',
'modified' => '2019-07-01 11:46:16',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 35 => array(
'id' => '3730',
'name' => 'Transcriptome-Wide Mapping 5-Methylcytosine by mC RNA Immunoprecipitation Followed by Deep Sequencing in Plant.',
'authors' => 'Gu X, Liang Z',
'description' => '<p>Transcriptome-wide mapping RNA modification is crucial to understand the distribution and function of RNA modifications. Here, we describe a protocol to transcriptome-wide mapping 5-methylcytosine (mC) in plant, by a RNA immunoprecipitation followed by deep sequencing (mC-RIP-seq) approach. The procedure includes RNA extraction, fragmentation, RNA immunoprecipitation, and library construction.</p>',
'date' => '2019-01-01',
'pmid' => 'http://www.pubmed.gov/30945199',
'doi' => '10.1007/978-1-4939-9045-0_24,',
'modified' => '2019-08-07 10:21:37',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 36 => array(
'id' => '3584',
'name' => 'Sensitivity of pituitary gonadotropes to hyperglycemia leads to epigenetic aberrations and reduced follicle-stimulating hormone levels.',
'authors' => 'Feldman A, Saleh A, Pnueli L, Qiao S, Shlomi T, Boehm U, Melamed P',
'description' => '<p>The connection between metabolism and reproductive function is well recognized, and we hypothesized that the pituitary gonadotropes, which produce luteinizing hormone and follicle-stimulating hormone (FSH), mediate some of the effects directly via insulin-independent glucose transporters, which allow continued glucose metabolism during hyperglycemia. We found that glucose transporter 1 is the predominant glucose transporter in primary gonadotropes and a gonadotrope precursor-derived cell line, and both are responsive to culture in high glucose; moreover, metabolite levels were altered in the cell line. Several of the affected metabolites are cofactors for chromatin-modifying enzymes, and in the gonadotrope precursor-derived cell line, we recorded global changes in histone acetylation and methylation, decreased DNA methylation, and increased hydroxymethylation, some of which did not revert to basal levels after cells were returned to normal glucose. Despite this weakening of epigenetic-mediated repression seen in the model cell line, FSH β-subunit ( Fshb) mRNA levels in primary gonadotropes were significantly reduced, apparently due in part to increased autocrine/paracrine effects of inhibin. However, unlike thioredoxin interacting protein and inhibin subunit α, Fshb mRNA levels did not recover after the return of cells to normal glucose. The effect on Fshb expression was also seen in 2 hyperglycemic mouse models, and levels of circulating FSH, required for follicle growth and development, were reduced. Thus, hyperglycemia seems to target the pituitary gonadotropes directly, and the likely extensive epigenetic changes are sensed acutely by Fshb. This scenario would explain clinical findings in which, even after restoration of optimal blood glucose levels, fertility often remains adversely affected. However, the relative accessibility of the pituitary provides a possible target for treatment, particularly crucial in the young in which hyperglycemia is increasingly common and fertility most relevant.-Feldman, A., Saleh, A., Pnueli, L., Qiao, S., Shlomi, T., Boehm, U., Melamed, P. Sensitivity of pituitary gonadotropes to hyperglycemia leads to epigenetic aberrations and reduced follicle-stimulating hormone levels.</p>',
'date' => '2018-12-27',
'pmid' => 'http://www.pubmed.gov/30074825',
'doi' => '10.1096/fj.201800943R',
'modified' => '2019-04-17 15:48:51',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 37 => array(
'id' => '3430',
'name' => 'Sensitive tumour detection and classification using plasma cell-free DNA methylomes.',
'authors' => 'Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, Kis O, Zhao Z, Spreafico A, Medina TDS, Wang Y, Roulois D, Ettayebi I, Chen Z, Chow S, Murphy T, Arruda A, O'Kane GM, Liu J, Mansour M, McPher',
'description' => '<p>The use of liquid biopsies for cancer detection and management is rapidly gaining prominence. Current methods for the detection of circulating tumour DNA involve sequencing somatic mutations using cell-free DNA, but the sensitivity of these methods may be low among patients with early-stage cancer given the limited number of recurrent mutations. By contrast, large-scale epigenetic alterations-which are tissue- and cancer-type specific-are not similarly constrained and therefore potentially have greater ability to detect and classify cancers in patients with early-stage disease. Here we develop a sensitive, immunoprecipitation-based protocol to analyse the methylome of small quantities of circulating cell-free DNA, and demonstrate the ability to detect large-scale DNA methylation changes that are enriched for tumour-specific patterns. We also demonstrate robust performance in cancer detection and classification across an extensive collection of plasma samples from several tumour types. This work sets the stage to establish biomarkers for the minimally invasive detection, interception and classification of early-stage cancers based on plasma cell-free DNA methylation patterns.</p>',
'date' => '2018-11-14',
'pmid' => 'http://www.pubmed.gov/30429608',
'doi' => '10.1038/s41586-018-0703-0',
'modified' => '2019-06-11 16:22:54',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 38 => array(
'id' => '3421',
'name' => 'Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate.',
'authors' => 'Sardina JL, Collombet S, Tian TV, Gómez A, Di Stefano B, Berenguer C, Brumbaugh J, Stadhouders R, Segura-Morales C, Gut M, Gut IG, Heath S, Aranda S, Di Croce L, Hochedlinger K, Thieffry D, Graf T',
'description' => '<p>Here, we report DNA methylation and hydroxymethylation dynamics at nucleotide resolution using C/EBPα-enhanced reprogramming of B cells into induced pluripotent cells (iPSCs). We observed successive waves of hydroxymethylation at enhancers, concomitant with a decrease in DNA methylation, suggesting active demethylation. Consistent with this finding, ablation of the DNA demethylase Tet2 almost completely abolishes reprogramming. C/EBPα, Klf4, and Tfcp2l1 each interact with Tet2 and recruit the enzyme to specific DNA sites. During reprogramming, some of these sites maintain high levels of 5hmC, and enhancers and promoters of key pluripotency factors become demethylated as early as 1 day after Yamanaka factor induction. Surprisingly, methylation changes precede chromatin opening in distinct chromatin regions, including Klf4 bound sites, revealing a pioneer factor activity associated with alternation in DNA methylation. Rapid changes in hydroxymethylation similar to those in B cells were also observed during compound-accelerated reprogramming of fibroblasts into iPSCs, highlighting the generality of our observations.</p>',
'date' => '2018-11-01',
'pmid' => 'http://www.pubmed.gov/30220521',
'doi' => '10.1016/j.stem.2018.08.016',
'modified' => '2018-12-31 11:16:24',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 39 => array(
'id' => '3409',
'name' => 'Oxidative stress in sperm affects the epigenetic reprogramming in early embryonic development.',
'authors' => 'Wyck S, Herrera C, Requena CE, Bittner L, Hajkova P, Bollwein H, Santoro R',
'description' => '<p>BACKGROUND: Reactive oxygen species (ROS)-induced oxidative stress is well known to play a major role in male infertility. Sperm are sensitive to ROS damaging effects because as male germ cells form mature sperm they progressively lose the ability to repair DNA damage. However, how oxidative DNA lesions in sperm affect early embryonic development remains elusive. RESULTS: Using cattle as model, we show that fertilization using sperm exposed to oxidative stress caused a major developmental arrest at the time of embryonic genome activation. The levels of DNA damage response did not directly correlate with the degree of developmental defects. The early cellular response for DNA damage, γH2AX, is already present at high levels in zygotes that progress normally in development and did not significantly increase at the paternal genome containing oxidative DNA lesions. Moreover, XRCC1, a factor implicated in the last step of base excision repair (BER) pathway, was recruited to the damaged paternal genome, indicating that the maternal BER machinery can repair these DNA lesions induced in sperm. Remarkably, the paternal genome with oxidative DNA lesions showed an impairment of zygotic active DNA demethylation, a process that previous studies linked to BER. Quantitative immunofluorescence analysis and ultrasensitive LC-MS-based measurements revealed that oxidative DNA lesions in sperm impair active DNA demethylation at paternal pronuclei, without affecting 5-hydroxymethylcytosine (5hmC), a 5-methylcytosine modification that has been implicated in paternal active DNA demethylation in mouse zygotes. Thus, other 5hmC-independent processes are implicated in active DNA demethylation in bovine embryos. The recruitment of XRCC1 to damaged paternal pronuclei indicates that oxidative DNA lesions drive BER to repair DNA at the expense of DNA demethylation. Finally, this study highlighted striking differences in DNA methylation dynamics between bovine and mouse zygotes that will facilitate the understanding of the dynamics of DNA methylation in early development. CONCLUSIONS: The data demonstrate that oxidative stress in sperm has an impact not only on DNA integrity but also on the dynamics of epigenetic reprogramming, which may harm the paternal genetic and epigenetic contribution to the developing embryo and affect embryo development and embryo quality.</p>',
'date' => '2018-10-17',
'pmid' => 'http://www.pubmed.gov/30333056',
'doi' => '10.1186/s13072-018-0224-y',
'modified' => '2018-11-09 11:10:58',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 40 => array(
'id' => '3404',
'name' => 'Integrated analysis of DNA methylation profiling and gene expression profiling identifies novel markers in lung cancer in Xuanwei, China.',
'authors' => 'Wang J, Duan Y, Meng QH, Gong R, Guo C, Zhao Y, Zhang Y',
'description' => '<p>BACKGROUND: Aberrant DNA methylation occurs frequently in cancer. The aim of this study was to identify novel methylation markers in lung cancer in Xuanwei, China, through integrated genome-wide DNA methylation and gene expression studies. METHODS: Differentially methylated regions (DMRs) and differentially expressed genes (DEGs) were detected on 10 paired lung cancer tissues and noncancerous lung tissues by methylated DNA immunoprecipitation combined with microarray (MeDIP-chip) and gene expression microarray analyses, respectively. Integrated analysis of DMRs and DEGs was performed to screen out candidate methylation-related genes. Both methylation and expression changes of the candidate genes were further validated and analyzed. RESULTS: Compared with normal lung tissues, lung cancer tissues expressed a total of 6,899 DMRs, including 5,788 hypermethylated regions and 1,111 hypomethylated regions. Integrated analysis of DMRs and DEGs identified 45 tumor-specific candidate genes: 38 genes whose DMRs were hypermethylated and expression was downregulated, and 7 genes whose DMRs were hypomethylated and expression was upregulated. The methylation and expression validation results identified 4 candidate genes (STXBP6, BCL6B, FZD10, and HSPB6) that were significantly hypermethylated and downregulated in most of the tumor tissues compared with the noncancerous lung tissues. CONCLUSIONS: This integrated analysis of genome-wide DNA methylation and gene expression in lung cancer in Xuanwei revealed several genes regulated by promoter methylation that have not been described in lung cancer before. These results provide new insight into the carcinogenesis of lung cancer in Xuanwei and represent promising new diagnostic and therapeutic targets.</p>',
'date' => '2018-10-04',
'pmid' => 'http://www.pubmed.gov/30286088',
'doi' => '10.1371/journal.pone.0203155',
'modified' => '2018-11-09 11:24:21',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 41 => array(
'id' => '3417',
'name' => 'mGlu1 Receptors Monopolize the Synaptic Control of Cerebellar Purkinje Cells by Epigenetically Down-Regulating mGlu5 Receptors.',
'authors' => 'Notartomaso S, Nakao H, Mascio G, Scarselli P, Cannella M, Zappulla C, Madonna M, Motolese M, Gradini R, Liberatore F, Zonta M, Carmignoto G, Battaglia G, Bruno V, Watanabe M, Aiba A, Nicoletti F',
'description' => '<p>In cerebellar Purkinje cells (PCs) type-1 metabotropic glutamate (mGlu1) receptors play a key role in motor learning and drive the refinement of synaptic innervation during postnatal development. The cognate mGlu5 receptor is absent in mature PCs and shows low expression levels in the adult cerebellar cortex. Here we found that mGlu5 receptors were heavily expressed by PCs in the early postnatal life, when mGlu1α receptors were barely detectable. The developmental decline of mGlu5 receptors coincided with the appearance of mGlu1α receptors in PCs, and both processes were associated with specular changes in CpG methylation in the corresponding gene promoters. It was the mGlu1 receptor that drove the elimination of mGlu5 receptors from PCs, as shown by data obtained with conditional mGlu1α receptor knockout mice and with targeted pharmacological treatments during critical developmental time windows. The suppressing activity of mGlu1 receptors on mGlu5 receptor was maintained in mature PCs, suggesting that expression of mGlu1α and mGlu5 receptors is mutually exclusive in PCs. These findings add complexity to the the finely tuned mechanisms that regulate PC biology during development and in the adult life and lay the groundwork for an in-depth analysis of the role played by mGlu5 receptors in PC maturation.</p>',
'date' => '2018-09-06',
'pmid' => 'http://www.pubmed.gov/30190524',
'doi' => '10.1038/s41598-018-31369-7',
'modified' => '2018-12-31 11:36:04',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 42 => array(
'id' => '3640',
'name' => 'Determination of the presence of 5-methylcytosine in Paramecium tetraurelia.',
'authors' => 'Singh A, Vancura A, Woycicki RK, Hogan DJ, Hendrick AG, Nowacki M',
'description' => '<p>5-methylcytosine DNA methylation regulates gene expression and developmental programming in a broad range of eukaryotes. However, its presence and potential roles in ciliates, complex single-celled eukaryotes with germline-somatic genome specialization via nuclear dimorphism, are largely uncharted. While canonical cytosine methyltransferases have not been discovered in published ciliate genomes, recent studies performed in the stichotrichous ciliate Oxytricha trifallax suggest de novo cytosine methylation during macronuclear development. In this study, we applied bisulfite genome sequencing, DNA mass spectrometry and antibody-based fluorescence detection to investigate the presence of DNA methylation in Paramecium tetraurelia. While the antibody-based methods suggest cytosine methylation, DNA mass spectrometry and bisulfite sequencing reveal that levels are actually below the limit of detection. Our results suggest that Paramecium does not utilize 5-methylcytosine DNA methylation as an integral part of its epigenetic arsenal.</p>',
'date' => '2018-06-07',
'pmid' => 'http://www.pubmed.gov/30379964',
'doi' => '10.1371/journal.',
'modified' => '2019-06-07 10:22:28',
'created' => '2019-06-06 12:11:18',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 43 => array(
'id' => '3458',
'name' => 'Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.',
'authors' => 'Li T, Wang L, Du Y, Xie S, Yang X, Lian F, Zhou Z, Qian C',
'description' => '<p>UHRF1 plays multiple roles in regulating DNMT1-mediated DNA methylation maintenance during DNA replication. The UHRF1 C-terminal RING finger functions as an ubiquitin E3 ligase to establish histone H3 ubiquitination at Lys18 and/or Lys23, which is subsequently recognized by DNMT1 to promote its localization onto replication foci. Here, we present the crystal structure of DNMT1 RFTS domain in complex with ubiquitin and highlight a unique ubiquitin binding mode for the RFTS domain. We provide evidence that UHRF1 N-terminal ubiquitin-like domain (UBL) also binds directly to DNMT1. Despite sharing a high degree of structural similarity, UHRF1 UBL and ubiquitin bind to DNMT1 in a very distinct fashion and exert different impacts on DNMT1 enzymatic activity. We further show that the UHRF1 UBL-mediated interaction between UHRF1 and DNMT1, and the binding of DNMT1 to ubiquitinated histone H3 that is catalyzed by UHRF1 RING domain are critical for the proper subnuclear localization of DNMT1 and maintenance of DNA methylation. Collectively, our study adds another layer of complexity to the regulatory mechanism of DNMT1 activation by UHRF1 and supports that individual domains of UHRF1 participate and act in concert to maintain DNA methylation patterns.</p>',
'date' => '2018-04-06',
'pmid' => 'http://www.pubmed.gov/29471350',
'doi' => '10.1093/nar/gky104',
'modified' => '2019-02-15 21:14:42',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 44 => array(
'id' => '3475',
'name' => 'Epigenetics and early domestication: differences in hypothalamic DNA methylation between red junglefowl divergently selected for high or low fear of humans.',
'authors' => 'Bélteky J, Agnvall B, Bektic L, Höglund A, Jensen P, Guerrero-Bosagna C',
'description' => '<p>BACKGROUND: Domestication of animals leads to large phenotypic alterations within a short evolutionary time-period. Such alterations are caused by genomic variations, yet the prevalence of modified traits is higher than expected if they were caused only by classical genetics and mutations. Epigenetic mechanisms may also be important in driving domesticated phenotypes such as behavior traits. Gene expression can be modulated epigenetically by mechanisms such as DNA methylation, resulting in modifications that are not only variable and susceptible to environmental stimuli, but also sometimes transgenerationally stable. To study such mechanisms in early domestication, we used as model two selected lines of red junglefowl (ancestors of modern chickens) that were bred for either high or low fear of humans over five generations, and investigated differences in hypothalamic DNA methylation between the two populations. RESULTS: Twenty-two 1-kb windows were differentially methylated between the two selected lines at p < 0.05 after false discovery rate correction. The annotated functions of the genes within these windows indicated epigenetic regulation of metabolic and signaling pathways, which agrees with the changes in gene expression that were previously reported for the same tissue and animals. CONCLUSIONS: Our results show that selection for an important domestication-related behavioral trait such as tameness can cause divergent epigenetic patterns within only five generations, and that these changes could have an important role in chicken domestication.</p>',
'date' => '2018-04-02',
'pmid' => 'http://www.pubmed.gov/29609558',
'doi' => '10.1186/s12711-018-0384-z',
'modified' => '2019-02-15 20:32:37',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 45 => array(
'id' => '3393',
'name' => 'Copper induces expression and methylation changes of early development genes in Crassostrea gigas embryos.',
'authors' => 'Sussarellu R, Lebreton M, Rouxel J, Akcha F, Rivière G',
'description' => '<p>Copper contamination is widespread along coastal areas and exerts adverse effects on marine organisms such as mollusks. In the Pacific oyster, copper induces severe developmental abnormalities during early life stages; however, the underlying molecular mechanisms are largely unknown. This study aims to better understand whether the embryotoxic effects of copper in Crassostrea gigas could be mediated by alterations in gene expression, and the putative role of DNA methylation, which is known to contribute to gene regulation in early embryo development. For that purpose, oyster embryos were exposed to 4 nominal copper concentrations (0.1, 1, 10 and 20 μg L Cu) during early development assays. Embryotoxicity was monitored through the oyster embryo-larval bioassay at the D-larva stage 24 h post fertilization (hpf) and genotoxicity at gastrulation 7 hpf. In parallel, the relative expression of 15 genes encoding putative homeotic, biomineralization and DNA methylation proteins was measured at three developmental stages (3 hpf morula stage, 7 hpf gastrula stage, 24 hpf D-larvae stage) using RT-qPCR. Global DNA content in methylcytosine and hydroxymethylcytosine were measured by HPLC and gene-specific DNA methylation levels were monitored using MeDIP-qPCR. A significant increase in larval abnormalities was observed from copper concentrations of 10 μg L, while significant genotoxic effects were detected at 1 μg L and above. All the selected genes presented a stage-dependent expression pattern, which was impaired for some homeobox and DNA methylation genes (Notochord, HOXA1, HOX2, Lox5, DNMT3b and CXXC-1) after copper exposure. While global DNA methylation (5-methylcytosine) at gastrula stage didn't show significant changes between experimental conditions, 5-hydroxymethylcytosine, its degradation product, decreased upon copper treatment. The DNA methylation of exons and the transcript levels were correlated in control samples for HOXA1 but such a correlation was diminished following copper exposure. The methylation level of some specific gene regions (HoxA1, Hox2, Engrailed2 and Notochord) displayed changes upon copper exposure. Such changes were gene and exon-specific and no obvious global trends could be identified. Our study suggests that the embryotoxic effects of copper in oysters could involve homeotic gene expression impairment possibly by changing DNA methylation levels.</p>',
'date' => '2018-03-01',
'pmid' => 'http://www.pubmed.gov/29353135',
'doi' => '10.1016/j.aquatox.2018.01.001',
'modified' => '2018-11-09 12:21:38',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 46 => array(
'id' => '3448',
'name' => 'Aberrant methylated key genes of methyl group metabolism within the molecular etiology of urothelial carcinogenesis.',
'authors' => 'Erichsen L, Ghanjati F, Beermann A, Poyet C, Hermanns T, Schulz WA, Seifert HH, Wild PJ, Buser L, Kröning A, Braunstein S, Anlauf M, Jankowiak S, Hassan M, Bendhack ML, Araúzo-Bravo MJ, Santourlidis S',
'description' => '<p>Urothelial carcinoma (UC), the most common cancer of the urinary bladder causes severe morbidity and mortality, e.g. about 40.000 deaths in the EU annually, and incurs considerable costs for the health system due to the need for prolonged treatments and long-term monitoring. Extensive aberrant DNA methylation is described to prevail in urothelial carcinoma and is thought to contribute to genetic instability, altered gene expression and tumor progression. However, it is unknown how this epigenetic alteration arises during carcinogenesis. Intact methyl group metabolism is required to ensure maintenance of cell-type specific methylomes and thereby genetic integrity and proper cellular function. Here, using two independent techniques for detecting DNA methylation, we observed DNA hypermethylation of the 5'-regulatory regions of the key methyl group metabolism genes ODC1, AHCY and MTHFR in early urothelial carcinoma. These hypermethylation events are associated with genome-wide DNA hypomethylation which is commonly associated with genetic instability. We therefore infer that hypermethylation of methyl group metabolism genes acts in a feed-forward cycle to promote additional DNA methylation changes and suggest a new hypothesis on the molecular etiology of urothelial carcinoma.</p>',
'date' => '2018-02-22',
'pmid' => 'http://www.pubmed.gov/29472622',
'doi' => '10.1038/s41598-018-21932-7',
'modified' => '2019-02-15 21:31:04',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 47 => array(
'id' => '3383',
'name' => 'Genome-wide analysis of day/night DNA methylation differences in Populus nigra.',
'authors' => 'Ding C.J. et al.',
'description' => '<p>DNA methylation is an important mechanism of epigenetic modification. Methylation changes during stress responses and developmental processes have been well studied; however, their role in plant adaptation to the day/night cycle is poorly understood. In this study, we detected global methylation patterns in leaves of the black poplar Populus nigra 'N46' at 8:00 and 24:00 by methylated DNA immunoprecipitation sequencing (MeDIP-seq). We found 10,027 and 10,242 genes to be methylated in the 8:00 and 24:00 samples, respectively. The methylated genes appeared to be involved in multiple biological processes, molecular functions, and cellular components, suggesting important roles for DNA methylation in poplar cells. Comparing the 8:00 and 24:00 samples, only 440 differentially methylated regions (DMRs) overlapped with genic regions, including 193 hyper- and 247 hypo-methylated DMRs, and may influence the expression of 137 downstream genes. Most hyper-methylated genes were associated with transferase activity, kinase activity, and phosphotransferase activity, whereas most hypo-methylated genes were associated with protein binding, ATP binding, and adenyl ribonucleotide binding, suggesting that different biological processes were activated during the day and night. Our results indicated that methylated genes were prevalent in the poplar genome, but that only a few of these participated in diurnal gene expression regulation.</p>',
'date' => '2018-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29293569',
'doi' => '',
'modified' => '2018-08-07 09:45:38',
'created' => '2018-08-07 09:45:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 48 => array(
'id' => '3384',
'name' => 'Obligatory and facilitative allelic variation in the DNA methylome within common disease-associated loci',
'authors' => 'Bell C.G. et al.',
'description' => '<p>Integrating epigenetic data with genome-wide association study (GWAS) results can reveal disease mechanisms. The genome sequence itself also shapes the epigenome, with CpG density and transcription factor binding sites (TFBSs) strongly encoding the DNA methylome. Therefore, genetic polymorphism impacts on the observed epigenome. Furthermore, large genetic variants alter epigenetic signal dosage. Here, we identify DNA methylation variability between GWAS-SNP risk and non-risk haplotypes. In three subsets comprising 3128 MeDIP-seq peripheral-blood DNA methylomes, we find 7173 consistent and functionally enriched Differentially Methylated Regions. 36.8% can be attributed to common non-SNP genetic variants. CpG-SNPs, as well as facilitative TFBS-motifs, are also enriched. Highlighting their functional potential, CpG-SNPs strongly associate with allele-specific DNase-I hypersensitivity sites. Our results demonstrate strong DNA methylation allelic differences driven by obligatory or facilitative genetic effects, with potential direct or regional disease-related repercussions. These allelic variations require disentangling from pure tissue-specific modifications, may influence array studies, and imply underestimated population variability in current reference epigenomes.</p>',
'date' => '2018-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29295990',
'doi' => '',
'modified' => '2018-08-07 10:13:12',
'created' => '2018-08-07 10:13:12',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 49 => array(
'id' => '3508',
'name' => 'Analysis of DNA methylome and transcriptome profiling following Gibberellin A3 (GA3) foliar application in Nicotiana tabacum L.',
'authors' => 'Manoharlal Raman, Saiprasad G. V. S., Kaikala Vinay, Suresh Kumar R., Kovařík Ales',
'description' => '<p>The present work investigated a comprehensive genome-wide landscape of DNA methylome and its relationship with transcriptome upon gibberellin A3 (GA3) foliar application under practical field conditions in solanaceae model, Nicotiana tabacum L. Methylated DNA Immunoprecipitation-Sequencing (MeDIP-Seq) analysis uncovered over 82% (18,456) of differential methylated regions (DMRs) in intergenic-region. Within protein-coding region, 2339 and 1685 of identified DMRs were observed in genebody- and promoter-region, respectively. Microarray study revealed 7032 differential expressed genes (DEGs) with 3507 and 3525 genes displaying upand down-regulation, respectively. Integration analysis revealed 520 unique non-redundant annotated DMRs overlapping with DEGs. Our results indicated that GA3 induced DNA hypo- as well as hyper-methylation were associated with both gene-silencing and -activation. No complete biasness or correlation was observed in either of the promoter- or genebody-regions, which otherwise showed an overall trend towards GA3 induced global DNA hypo-methylation. Taken together, our results suggested that differential DNA methylation mediated by GA3 may only play a permissive role in regulating the gene expression.</p>',
'date' => '2018-01-01',
'pmid' => 'https://link.springer.com/article/10.1007/s40502-018-0393-5',
'doi' => '10.1007/s40502-018-0393-5',
'modified' => '2022-05-18 18:43:47',
'created' => '2019-02-27 12:54:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 50 => array(
'id' => '3297',
'name' => '5-Methylcytosine RNA Methylation in Arabidopsis Thaliana',
'authors' => 'Cui X. et al.',
'description' => '<p>5-Methylcytosine (m<sup>5</sup>C) is a well-characterized DNA modification, and is also predominantly reported in abundant non-coding RNAs in both prokaryotes and eukaryotes. However, the distribution and biological functions of m<sup>5</sup>C in plant mRNAs remain largely unknown. Here, we report transcriptome-wide profiling of RNA m<sup>5</sup>C in Arabidopsis thaliana by applying m<sup>5</sup>C RNA immunoprecipitation followed by a deep-sequencing approach (m<sup>5</sup>C-RIP-seq). LC-MS/MS and dot blot analyses reveal a dynamic pattern of m<sup>5</sup>C mRNA modification in various tissues and at different developmental stages. m<sup>5</sup>C-RIP-seq analysis identified 6045 m<sup>5</sup>C peaks in 4465 expressed genes in young seedlings. We found that m<sup>5</sup>C is enriched in coding sequences with two peaks located immediately after start codons and before stop codons, and is associated with mRNAs with low translation activity. We further demonstrated that an RNA (cytosine-5)-methyltransferase, tRNA-specific methyltransferase 4B (TRM4B), exhibits m<sup>5</sup>C RNA methyltransferase activity. Mutations in TRM4B display defects in root development and decreased m<sup>5</sup>C peaks. TRM4B affects the transcript levels of the genes involved in root development, which is positively correlated with their mRNA stability and m<sup>5</sup>C levels. Our results suggest that m<sup>5</sup>C in mRNA is a new epitranscriptome marker inArabidopsis, and that regulation of this modification is an integral part of gene regulatory networks underlying plant development.</p>',
'date' => '2017-11-06',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28965832',
'doi' => '',
'modified' => '2017-12-04 11:10:34',
'created' => '2017-12-04 11:10:34',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 51 => array(
'id' => '3220',
'name' => 'Maternal obesity programs increased leptin gene expression in rat male offspring via epigenetic modifications in a depot-specific manner',
'authors' => 'Lecoutre S. et al.',
'description' => '<div class="">
<h4>OBJECTIVE:</h4>
<p><abstracttext label="OBJECTIVE" nlmcategory="OBJECTIVE">According to the Developmental Origin of Health and Disease (DOHaD) concept, maternal obesity and accelerated growth in neonates predispose offspring to white adipose tissue (WAT) accumulation. In rodents, adipogenesis mainly develops during lactation. The mechanisms underlying the phenomenon known as developmental programming remain elusive. We previously reported that adult rat offspring from high-fat diet-fed dams (called HF) exhibited hypertrophic adipocyte, hyperleptinemia and increased leptin mRNA levels in a depot-specific manner. We hypothesized that leptin upregulation occurs via epigenetic malprogramming, which takes place early during development of WAT.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">As a first step, we identified <i>in silico</i> two potential enhancers located upstream and downstream of the leptin transcription start site that exhibit strong dynamic epigenomic remodeling during adipocyte differentiation. We then focused on epigenetic modifications (methylation, hydroxymethylation, and histone modifications) of the promoter and the two potential enhancers regulating leptin gene expression in perirenal (pWAT) and inguinal (iWAT) fat pads of HF offspring during lactation (postnatal days 12 (PND12) and 21 (PND21)) and in adulthood.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">PND12 is an active period for epigenomic remodeling in both deposits especially in the upstream enhancer, consistent with leptin gene induction during adipogenesis. Unlike iWAT, some of these epigenetic marks were still observable in pWAT of weaned HF offspring. Retained marks were only visible in pWAT of 9-month-old HF rats that showed a persistent "expandable" phenotype.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Consistent with the DOHaD hypothesis, persistent epigenetic remodeling occurs at regulatory regions especially within intergenic sequences, linked to higher leptin gene expression in adult HF offspring in a depot-specific manner.</abstracttext></p>
</div>',
'date' => '2017-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5518658/',
'doi' => '',
'modified' => '2017-08-18 13:56:40',
'created' => '2017-08-18 13:56:40',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 52 => array(
'id' => '3204',
'name' => 'Increased 5-hydroxymethylation levels in the hippocampus of rat extinguished from cocaine self-administration',
'authors' => 'Sadakierska-Chudy A. et al.',
'description' => '<p>Drug craving and relapse risk during abstinence from cocaine are thought to be caused by persistent changes in transcription and chromatin regulation. Although several brain regions are involved in these processes, the hippocampus seems to play an important role in context-evoked craving and drug-seeking behavior. Only a few studies have examined epigenetic alterations during a period of cocaine abstinence. To investigate the effects of cocaine abstinence on DNA methylation and gene expression, rats that self-administered the drug underwent cocaine abstinence in two time points with extinction training. During the cocaine extinction, we observed elevated global 5-hydroxymethylcytosine(5-hmC) levels with a concurrent increase in Tet3 transcript levels. Moreover, we did not find significant alterations in the levels of Tet3 mRNA and 5-hmC in rats subjected to cocaine abstinence without extinction training. Additionally, our findings demonstrated that the expression of Tet3 target genes was activated. Besides, altered DNA methylation was detected at promoter regions of miRNAs, such as miR-30d and miR-let7i. Further in silico analysis provided evidence that these two molecules targeted the 3' UTR region of the Tet3 gene and thus may contribute to its post-transcriptional regulation. This study has presented novel findings in the hippocampus of rats that underwent extinction training following cocaine self-administration. The alterations in the Tet3 gene expression and the level of 5-hmC may play an important role in extinction learning and the reduction of subsequent cocaine seeking.</p>',
'date' => '2017-07-27',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28422379',
'doi' => '',
'modified' => '2017-07-03 10:21:48',
'created' => '2017-07-03 10:21:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 53 => array(
'id' => '3225',
'name' => 'The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation',
'authors' => 'Leone S. et al.',
'description' => '<p>Long non-coding RNAs (lncRNAs) have been implicated in the regulation of chromatin conformation and epigenetic patterns. lncRNA expression levels are widely taken as an indicator for functional properties. However, the role of RNA processing in modulating distinct features of the same lncRNA is less understood. The establishment of heterochromatin at rRNA genes depends on the processing of IGS-rRNA into pRNA, a reaction that is impaired in embryonic stem cells (ESCs) and activated only upon differentiation. The production of mature pRNA is essential since it guides the repressor TIP5 to rRNA genes, and IGS-rRNA abolishes this process. Through screening for IGS-rRNA-binding proteins, we here identify the RNA helicase DHX9 as a regulator of pRNA processing. DHX9 binds to rRNA genes only upon ESC differentiation and its activity guides TIP5 to rRNA genes and establishes heterochromatin. Remarkably, ESCs depleted of DHX9 are unable to differentiate and this phenotype is reverted by the addition of pRNA, whereas providing IGS-rRNA and pRNA mutants deficient for TIP5 binding are not sufficient. Our results reveal insights into lncRNA biogenesis during development and support a model in which the state of rRNA gene chromatin is part of the regulatory network that controls exit from pluripotency and initiation of differentiation pathways.</p>',
'date' => '2017-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28588071',
'doi' => '',
'modified' => '2017-08-22 13:52:28',
'created' => '2017-08-22 13:52:28',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 54 => array(
'id' => '3233',
'name' => 'Pramel7 mediates ground-state pluripotency through proteasomal-epigenetic combined pathways.',
'authors' => 'Graf U. et al.',
'description' => '<p>Naive pluripotency is established in preimplantation epiblast. Embryonic stem cells (ESCs) represent the immortalization of naive pluripotency. 2i culture has optimized this state, leading to a gene signature and DNA hypomethylation closely comparable to preimplantation epiblast, the developmental ground state. Here we show that Pramel7 (PRAME-like 7), a protein highly expressed in the inner cell mass (ICM) but expressed at low levels in ESCs, targets for proteasomal degradation UHRF1, a key factor for DNA methylation maintenance. Increasing Pramel7 expression in serum-cultured ESCs promotes a preimplantation epiblast-like gene signature, reduces UHRF1 levels and causes global DNA hypomethylation. Pramel7 is required for blastocyst formation and its forced expression locks ESCs in pluripotency. Pramel7/UHRF1 expression is mutually exclusive in ICMs whereas Pramel7-knockout embryos express high levels of UHRF1. Our data reveal an as-yet-unappreciated dynamic nature of DNA methylation through proteasome pathways and offer insights that might help to improve ESC culture to reproduce in vitro the in vivo ground-state pluripotency.</p>',
'date' => '2017-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28604677',
'doi' => '',
'modified' => '2017-08-24 09:57:28',
'created' => '2017-08-24 09:57:28',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 55 => array(
'id' => '3200',
'name' => 'CHD4 Has Oncogenic Functions in Initiating and Maintaining Epigenetic Suppression of Multiple Tumor Suppressor Genes',
'authors' => 'Xia L. et al.',
'description' => '<p>An oncogenic role for CHD4, a NuRD component, is defined for initiating and supporting tumor suppressor gene (TSG) silencing in human colorectal cancer. CHD4 recruits repressive chromatin proteins to sites of DNA damage repair, including DNA methyltransferases where it imposes de novo DNA methylation. At TSGs, CHD4 retention helps maintain DNA hypermethylation-associated transcriptional silencing. CHD4 is recruited by the excision repair protein OGG1 for oxidative damage to interact with the damage-induced base 8-hydroxydeoxyguanosine (8-OHdG), while ZMYND8 recruits it to double-strand breaks. CHD4 knockdown activates silenced TSGs, revealing their role for blunting colorectal cancer cell proliferation, invasion, and metastases. High CHD4 and 8-OHdG levels plus low expression of TSGs strongly correlates with early disease recurrence and decreased overall survival.</p>',
'date' => '2017-05-08',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28486105',
'doi' => '',
'modified' => '2017-07-03 09:56:32',
'created' => '2017-07-03 09:56:32',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 56 => array(
'id' => '3170',
'name' => 'Critical threshold levels of DNA methyltransferase 1 are required to maintain DNA methylation across the genome in human cancer cells.',
'authors' => 'Cai Y. et al.',
'description' => '<p>Reversing DNA methylation abnormalities and associated gene silencing, through inhibiting DNA methyltransferases (DNMTs) is an important potential cancer therapy paradigm. Maximizing this potential requires defining precisely how these enzymes maintain genome-wide, cancer-specific DNA methylation. To date, there is incomplete understanding of precisely how the three DNMTs, 1, 3A, and 3B, interact for maintaining DNA methylation abnormalities in cancer. By combining genetic and shRNA depletion strategies, we define not only a dominant role for DNA methyltransferase 1 (DNMT1) but also distinct roles of 3A and 3B in genome-wide DNA methylation maintenance. Lowering DNMT1 below a threshold level is required for maximal loss of DNA methylation at all genomic regions, including gene body and enhancer regions, and for maximally reversing abnormal promoter DNA hypermethylation and associated gene silencing to reexpress key genes. It is difficult to reach this threshold with patient-tolerable doses of current DNMT inhibitors (DNMTIs). We show that new approaches, like decreasing the DNMT targeting protein, UHRF1, can augment the DNA demethylation capacities of existing DNA methylation inhibitors for fully realizing their therapeutic potential.</p>',
'date' => '2017-04-27',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28232479',
'doi' => '',
'modified' => '2017-05-10 15:31:12',
'created' => '2017-05-10 15:31:12',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 57 => array(
'id' => '3228',
'name' => 'Regulation of DNA demethylation by the XPC DNA repair complex in somatic and pluripotent stem cells.',
'authors' => 'Ho J.J. et al.',
'description' => '<p>Faithful resetting of the epigenetic memory of a somatic cell to a pluripotent state during cellular reprogramming requires DNA methylation to silence somatic gene expression and dynamic DNA demethylation to activate pluripotency gene transcription. The removal of methylated cytosines requires the base excision repair enzyme TDG, but the mechanism by which TDG-dependent DNA demethylation occurs in a rapid and site-specific manner remains unclear. Here we show that the XPC DNA repair complex is a potent accelerator of global and locus-specific DNA demethylation in somatic and pluripotent stem cells. XPC cooperates with TDG genome-wide to stimulate the turnover of essential intermediates by overcoming slow TDG-abasic product dissociation during active DNA demethylation. We further establish that DNA demethylation induced by XPC expression in somatic cells overcomes an early epigenetic barrier in cellular reprogramming and facilitates the generation of more robust induced pluripotent stem cells, characterized by enhanced pluripotency-associated gene expression and self-renewal capacity. Taken together with our previous studies establishing the XPC complex as a transcriptional coactivator, our findings underscore two distinct but complementary mechanisms by which XPC influences gene regulation by coordinating efficient TDG-mediated DNA demethylation along with active transcription during somatic cell reprogramming.</p>',
'date' => '2017-04-05',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28512237',
'doi' => '',
'modified' => '2017-08-23 14:20:13',
'created' => '2017-08-23 14:20:13',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 58 => array(
'id' => '3142',
'name' => 'Epigenetic regulation of RELN and GAD1 in the frontal cortex (FC) of autism spectrum disorder (ASD) subjects',
'authors' => 'Zhubi A. et al.',
'description' => '<p>Both Reelin (RELN) and glutamate decarboxylase 67 (GAD1) have been implicated in the pathophysiology of Autism Spectrum Disorders (ASD). We have previously shown that both mRNAs are reduced in the cerebella (CB) of ASD subjects through a mechanism that involves increases in the amounts of MECP2 binding to the corresponding promoters. In the current study, we examined the expression of RELN, GAD1, GAD2, and several other mRNAs implicated in this disorder in the frontal cortices (FC) of ASD and CON subjects. We also focused on the role that epigenetic processes play in the regulation of these genes in ASD brain. Our goal is to better understand the molecular basis for the down-regulation of genes expressed in GABAergic neurons in ASD brains. We measured mRNA levels corresponding to selected GABAergic genes using qRT-PCR in RNA isolated from both ASD and CON groups. We determined the extent of binding of MECP2 and DNMT1 repressor proteins by chromatin immunoprecipitation (ChIP) assays. The amount of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) present in the promoters of the target genes was quantified by methyl DNA immunoprecipitation (MeDIP) and hydroxyl MeDIP (hMeDIP). We detected significant reductions in the mRNAs associated with RELN and GAD1 and significant increases in mRNAs encoding the Ten-eleven Translocation (TET) enzymes 1, 2, and 3. We also detected increased MECP2 and DNMT1 binding to the corresponding promoter regions of GAD1, RELN, and GAD2. Interestingly, there were decreased amounts of 5mC at both promoters and little change in 5hmC content in these same DNA fragments. Our data demonstrate that RELN, GAD1, and several other genes selectively expressed in GABAergic neurons, are down-regulated in post-mortem ASD FC. In addition, we observed increased DNMT1 and MECP2 binding at the corresponding promoters of these genes. The finding of increased MECP2 binding to the RELN, GAD1 and GAD2 promoters, with reduced amounts of 5mC and unchanged amounts of 5hmC present in these regions, suggests the possibility that DNMT1 interacts with and alters MECP2 binding properties to selected promoters. Comparisons between data obtained from the FC with CB studies showed some common themes between brain regions which are discussed.</p>',
'date' => '2017-02-14',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28229923',
'doi' => '',
'modified' => '2017-03-23 14:58:21',
'created' => '2017-03-23 14:58:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 59 => array(
'id' => '3126',
'name' => 'Intergenerational Transmission of Enhanced Seizure Susceptibility after Febrile Seizures',
'authors' => 'Wu D. et al.',
'description' => '<p>Environmental exposure early in development plays a role in susceptibility to disease in later life. Here, we demonstrate that prolonged febrile seizures induced by exposure of rat pups to a hyperthermic environment enhance seizure susceptibility not only in these hyperthermia-treated rats but also in their future offspring, even if the offspring never experience febrile seizures. This transgenerational transmission was intensity-dependent and was mainly from mothers to their offspring. The transmission was associated with DNA methylation. Thus, our study supports a “Lamarckian”-like mechanism of pathogenesis and the crucial role of epigenetic factors in neurological conditions.</p>',
'date' => '2017-02-08',
'pmid' => 'http://www.sciencedirect.com/science/article/pii/S2352396417300658',
'doi' => '',
'modified' => '2017-02-23 11:05:25',
'created' => '2017-02-23 11:05:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 60 => array(
'id' => '3125',
'name' => 'Pharmacological inhibition of DNA methyltransferase 1 promotes neuronal differentiation from rodent and human nasal olfactory stem/progenitor cell cultures',
'authors' => 'Franco I. et al.',
'description' => '<p>Nasal olfactory stem and neural progenitor cells (NOS/PCs) are considered possible tools for regenerative stem cell therapies in neurodegenerative diseases. Neurogenesis is a complex process regulated by extrinsic and intrinsic signals that include DNA-methylation and other chromatin modifications that could be experimentally manipulated in order to increase neuronal differentiation. The aim of the present study was the characterization of primary cultures and consecutive passages (P2-P10) of NOS/PCs isolated from male Swiss-Webster (mNOS/PCs) or healthy humans (hNOS/PCs). We evaluated and compared cellular morphology, proliferation rates and the expression pattern of pluripotency-associated markers and DNA methylation-associated gene expression in these cultures. Neuronal differentiation was induced by exposure to all-trans retinoic acid and forskolin for 7 days and evaluated by morphological analysis and immunofluorescence against neuronal markers MAP2, NSE and MAP1B. In response to the inductive cues mNOS/PCs expressed NSE (75.67%) and MAP2 (35.34%); whereas the majority of the hNOS/PCs were immunopositive to MAP1B. Treatment with procainamide, a specific inhibitor of DNA methyltransferase 1 (DNMT1), increases in the number of forskolin‘/retinoic acid-induced mature neuronal marker-expressing mNOS/PCs cells and enhances neurite development in hNOS/PCs. Our results indicate that mice and human nasal olfactory stem/progenitors cells share pluripotency-related gene expression suggesting that their application for stem cell therapy is worth pursuing and that DNA methylation inhibitors could be efficient tools to enhance neuronal differentiation from these cells.</p>',
'date' => '2017-02-01',
'pmid' => 'http://www.sciencedirect.com/science/article/pii/S0736574816303665',
'doi' => '',
'modified' => '2017-02-16 10:34:07',
'created' => '2017-02-16 10:34:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 61 => array(
'id' => '3119',
'name' => 'Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain',
'authors' => 'Amort T. et al.',
'description' => '<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec1">
<h3 xmlns="" class="Heading">Background</h3>
<p id="Par1" class="Para">Recent work has identified and mapped a range of posttranscriptional modifications in mRNA, including methylation of the N6 and N1 positions in adenine, pseudouridylation, and methylation of carbon 5 in cytosine (m5C). However, knowledge about the prevalence and transcriptome-wide distribution of m5C is still extremely limited; thus, studies in different cell types, tissues, and organisms are needed to gain insight into possible functions of this modification and implications for other regulatory processes.</p>
</div>
<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec2">
<h3 xmlns="" class="Heading">Results</h3>
<p id="Par2" class="Para">We have carried out an unbiased global analysis of m5C in total and nuclear poly(A) RNA of mouse embryonic stem cells and murine brain. We show that there are intriguing differences in these samples and cell compartments with respect to the degree of methylation, functional classification of methylated transcripts, and position bias within the transcript. Specifically, we observe a pronounced accumulation of m5C sites in the vicinity of the translational start codon, depletion in coding sequences, and mixed patterns of enrichment in the 3′ UTR. Degree and pattern of methylation distinguish transcripts modified in both embryonic stem cells and brain from those methylated in either one of the samples. We also analyze potential correlations between m5C and micro RNA target sites, binding sites of RNA binding proteins, and <em xmlns="" class="EmphasisTypeItalic">N</em>6-methyladenosine.</p>
</div>
<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec3">
<h3 xmlns="" class="Heading">Conclusion</h3>
<p id="Par3" class="Para">Our study presents the first comprehensive picture of cytosine methylation in the epitranscriptome of pluripotent and differentiated stages in the mouse. These data provide an invaluable resource for future studies of function and biological significance of m5C in mRNA in mammals.</p>
</div>',
'date' => '2017-01-05',
'pmid' => 'https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1139-1',
'doi' => '',
'modified' => '2017-02-14 17:20:08',
'created' => '2017-02-14 17:20:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 62 => array(
'id' => '3061',
'name' => 'Novel regional age-associated DNA methylation changes within human common disease-associated loci',
'authors' => 'Bell CG et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">Advancing age progressively impacts on risk and severity of chronic disease. It also modifies the epigenome, with changes in DNA methylation, due to both random drift and variation within specific functional loci.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">In a discovery set of 2238 peripheral-blood genome-wide DNA methylomes aged 19-82 years, we identify 71 age-associated differentially methylated regions within the linkage disequilibrium blocks of the single nucleotide polymorphisms from the NIH genome-wide association study catalogue. This included 52 novel regions, 29 within loci not covered by 450 k or 27 k Illumina array, and with enrichment for DNase-I Hypersensitivity sites across the full range of tissues. These age-associated differentially methylated regions also show marked enrichment for enhancers and poised promoters across multiple cell types. In a replication set of 2084 DNA methylomes, 95.7 % of the age-associated differentially methylated regions showed the same direction of ageing effect, with 80.3 % and 53.5 % replicated to p < 0.05 and p < 1.85 × 10<sup>-8</sup>, respectively.</abstracttext></p>
<h4>CONCLUSION:</h4>
<p><abstracttext label="CONCLUSION" nlmcategory="CONCLUSIONS">By analysing the functionally enriched disease and trait-associated regions of the human genome, we identify novel epigenetic ageing changes, which could be useful biomarkers or provide mechanistic insights into age-related common diseases.</abstracttext></p>
</div>',
'date' => '2016-09-26',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27663977',
'doi' => '',
'modified' => '2016-11-04 10:56:10',
'created' => '2016-11-02 09:54:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 63 => array(
'id' => '3007',
'name' => '5-hydroxymethylcytosine marks postmitotic neural cells in the adult and developing vertebrate central nervous system',
'authors' => 'Diotel N et al.',
'description' => '<p>The epigenetic mark 5-hydroxymethylcytosine (5hmC) is a cytosine modification that is abundant in the central nervous system of mammals and which results from 5-methylcytosine oxidation by TET enzymes. Such a mark is suggested to play key roles in the regulation of chromatin structure and gene expression. However, its precise functions still remain poorly understood and information about its distribution in non-mammalian species is still lacking. Here, the distribution of 5hmC was investigated in the brain of adult zebrafish, African claw frog, and mouse in a comparative manner. We show that zebrafish neurons are endowed with high levels of 5hmC, whereas quiescent or proliferative neural progenitors show low to undetectable levels of the modified cytosine. In the brain of larval and juvenile Xenopus, 5hmC is also detected in neurons, while ventricular proliferative cells do not display this epigenetic mark. Similarly, 5hmC is enriched in neurons compared to neural progenitors of the ventricular zone in the mouse developing cortex. Interestingly, 5hmC colocalized with the methylated DNA binding protein MeCP2 and with the active chromatin histone modification H3K4me2 in mouse neurons. Taken together, our results show an evolutionarily conserved cerebral distribution of 5hmC between fish and tetrapods and reinforce the idea that 5hmC fulfills major functions in the control of chromatin activity in vertebrate neurons.</p>',
'date' => '2016-07-14',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27414756',
'doi' => '',
'modified' => '2016-08-29 09:24:44',
'created' => '2016-08-29 09:24:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 64 => array(
'id' => '2992',
'name' => 'Regulation of the DNA Methylation Landscape in Human Somatic Cell Reprogramming by the miR-29 Family',
'authors' => 'Hysolli E et al.',
'description' => 'Reprogramming to pluripotency after overexpression of OCT4, SOX2, KLF4, and MYC is accompanied by global genomic and epigenomic changes. Histone modification and DNA methylation states in induced pluripotent stem cells (iPSCs) have been shown to be highly similar to embryonic stem cells (ESCs). However, epigenetic differences still exist between iPSCs and ESCs. In particular, aberrant DNA methylation states found in iPSCs are a major concern when using iPSCs in a clinical setting. Thus, it is critical to find factors that regulate DNA methylation states in reprogramming. Here, we found that the miR-29 family is an important epigenetic regulator during human somatic cell reprogramming. Our global DNA methylation and hydroxymethylation analysis shows that DNA demethylation is a major event mediated by miR-29a depletion during early reprogramming, and that iPSCs derived from miR-29a depletion are epigenetically closer to ESCs. Our findings uncover an important miRNA-based approach to generate clinically robust iPSCs.',
'date' => '2016-07-12',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27373925',
'doi' => '10.1016/j.stemcr.2016.05.014',
'modified' => '2016-08-23 09:57:29',
'created' => '2016-08-23 09:57:29',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 65 => array(
'id' => '3011',
'name' => 'Genome-Wide DNA Methylation in Mixed Ancestry Individuals with Diabetes and Prediabetes from South Africa',
'authors' => 'Matsha TE et al.',
'description' => '<p>Aims. To conduct a genome-wide DNA methylation in individuals with type 2 diabetes, individuals with prediabetes, and control mixed ancestry individuals from South Africa. Methods. We used peripheral blood to perform genome-wide DNA methylation analysis in 3 individuals with screen detected diabetes, 3 individuals with prediabetes, and 3 individuals with normoglycaemia from the Bellville South Community, Cape Town, South Africa, who were age-, gender-, body mass index-, and duration of residency-matched. Methylated DNA immunoprecipitation (MeDIP) was performed by Arraystar Inc. (Rockville, MD, USA). Results. Hypermethylated DMRs were 1160 (81.97%) and 124 (43.20%), respectively, in individuals with diabetes and prediabetes when both were compared to subjects with normoglycaemia. Our data shows that genes related to the immune system, signal transduction, glucose transport, and pancreas development have altered DNA methylation in subjects with prediabetes and diabetes. Pathway analysis based on the functional analysis mapping of genes to KEGG pathways suggested that the linoleic acid metabolism and arachidonic acid metabolism pathways are hypomethylated in prediabetes and diabetes. Conclusions. Our study suggests that epigenetic changes are likely to be an early process that occurs before the onset of overt diabetes. Detailed analysis of DMRs that shows gradual methylation differences from control versus prediabetes to prediabetes versus diabetes in a larger sample size is required to confirm these findings.</p>',
'date' => '2016-06-28',
'pmid' => 'http://www.hindawi.com/journals/ije/2016/3172093/',
'doi' => '',
'modified' => '2016-08-29 10:27:14',
'created' => '2016-08-29 10:27:14',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 66 => array(
'id' => '2954',
'name' => 'Dnmt2/Trdmt1 as Mediator of RNA Polymerase II Transcriptional Activity in Cardiac Growth',
'authors' => 'Ghanbarian H et al.',
'description' => '<p>Dnmt2/Trdmt1 is a methyltransferase, which has been shown to methylate tRNAs. Deficient mutants were reported to exhibit various, seemingly unrelated, defects in development and RNA-mediated epigenetic heredity. Here we report a role in a distinct developmental regulation effected by a noncoding RNA. We show that Dnmt2-deficiency in mice results in cardiac hypertrophy. Echocardiographic measurements revealed that cardiac function is preserved notwithstanding the increased dimensions of the organ due to cardiomyocyte enlargement. Mechanistically, activation of the P-TEFb complex, a critical step for cardiac growth, results from increased dissociation of the negatively regulating Rn7sk non-coding RNA component in Dnmt2-deficient cells. Our data suggest that Dnmt2 plays an unexpected role for regulation of cardiac growth by modulating activity of the P-TEFb complex.</p>',
'date' => '2016-06-06',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27270731',
'doi' => ' 10.1371/journal.pone.0156953',
'modified' => '2016-06-14 15:49:17',
'created' => '2016-06-14 15:49:17',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 67 => array(
'id' => '2941',
'name' => 'Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers',
'authors' => 'Li L et al.',
'description' => '<p>Promoter CpG methylation is a fundamental regulatory process of gene expression. TET proteins are active CpG demethylases converting 5-methylcytosine to 5-hydroxymethylcytosine, with loss of 5 hmC as an epigenetic hallmark of cancers, indicating critical roles of TET proteins in epigenetic tumorigenesis. Through analysis of tumor methylomes, we discovered <i>TET1</i> as a methylated target, and further confirmed its frequent downregulation/methylation in cell lines and primary tumors of multiple carcinomas and lymphomas, including nasopharyngeal, esophageal, gastric, colorectal, renal, breast and cervical carcinomas, as well as non-Hodgkin, Hodgkin and nasal natural killer/T-cell lymphomas, although all three <i>TET</i> family genes are ubiquitously expressed in normal tissues. Ectopic expression of TET1 catalytic domain suppressed colony formation and induced apoptosis of tumor cells of multiple tissue types, supporting its role as a broad <i>bona fide</i> tumor suppressor. Furthermore, TET1 catalytic domain possessed demethylase activity in cancer cells, being able to inhibit the CpG methylation of tumor suppressor gene (TSG) promoters and reactivate their expression, such as <i>SLIT2, ZNF382</i> and <i>HOXA9</i>. As only infrequent mutations of <i>TET1</i> have been reported, compared to <i>TET2</i>, epigenetic silencing therefore appears to be the dominant mechanism for <i>TET1</i> inactivation in cancers, which also forms a feedback loop of CpG methylation during tumorigenesis.</p>',
'date' => '2016-05-26',
'pmid' => 'http://www.nature.com/articles/srep26591',
'doi' => '10.1038/srep26591',
'modified' => '2016-06-06 09:47:31',
'created' => '2016-06-06 09:47:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 68 => array(
'id' => '2836',
'name' => 'Biochemical reconstitution of TET1–TDG–BER-dependent active DNA demethylation reveals a highly coordinated mechanism',
'authors' => 'Weber AR, Krawczyk C, Robertson AB, Kuśnierczyk A, Vågbø CB, Schuermann D, Klungland A, Schär P',
'description' => '<p>Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten–eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET–TDG–BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.nature.com/ncomms/2016/160302/ncomms10806/full/ncomms10806.html',
'doi' => '10.1038/ncomms10806',
'modified' => '2016-03-08 10:25:46',
'created' => '2016-03-08 10:25:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 69 => array(
'id' => '2837',
'name' => 'Hydroxymethylation of microRNA-365-3p Regulates Nociceptive Behaviors via Kcnh2',
'authors' => 'Pan Z, Zhang M, Ma T, Xue Z-Y, Li G-F, Hao L-Y, Zhu L-J, Li Y-Q, Ding H-L, Cao J-L',
'description' => '<p id="p-3">DNA 5-hydroxylmethylcytosine (5hmC) catalyzed by ten-eleven translocation methylcytosine dioxygenase (TET) occurs abundantly in neurons of mammals. However, the <em>in vivo</em> causal link between TET dysregulation and nociceptive modulation has not been established. Here, we found that spinal TET1 and TET3 were significantly increased in the model of formalin-induced acute inflammatory pain, which was accompanied with the augment of genome-wide 5hmC content in spinal cord. Knockdown of spinal TET1 or TET3 alleviated the formalin-induced nociceptive behavior and overexpression of spinal TET1 or TET3 in naive mice produced pain-like behavior as evidenced by decreased thermal pain threshold. Furthermore, we found that TET1 or TET3 regulated the nociceptive behavior by targeting microRNA-365-3p (miR-365-3p). Formalin increased 5hmC in the miR-365-3p promoter, which was inhibited by knockdown of TET1 or TET3 and mimicked by overexpression of TET1 or TET3 in naive mice. Nociceptive behavior induced by formalin or overexpression of spinal TET1 or TET3 could be prevented by downregulation of miR-365-3p, and mimicked by overexpression of spinal miR-365-3p. Finally, we demonstrated that a potassium channel, voltage-gated eag-related subfamily H member 2 (<em>Kcnh2</em>), validated as a target of miR-365-3p, played a critical role in nociceptive modulation by spinal TET or miR-365-3p. Together, we concluded that TET-mediated hydroxymethylation of miR-365-3p regulates nociceptive behavior via <em>Kcnh2</em>.</p>
<p id="p-4"><strong>SIGNIFICANCE STATEMENT</strong> Mounting evidence indicates that epigenetic modifications in the nociceptive pathway contribute to pain processes and analgesia response. Here, we found that the increase of 5hmC content mediated by TET1 or TET3 in miR-365-3p promoter in the spinal cord is involved in nociceptive modulation through targeting a potassium channel, <em>Kcnh2</em>. Our study reveals a new epigenetic mechanism underlying nociceptive information processing, which may be a novel target for development of antinociceptive drugs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.jneurosci.org/content/36/9/2769.short',
'doi' => '10.1523/JNEUROSCI.3474-15.2016',
'modified' => '2016-03-08 10:40:48',
'created' => '2016-03-08 10:40:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 70 => array(
'id' => '3034',
'name' => 'Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism',
'authors' => 'Weber AR et al.',
'description' => '<p>Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten-eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET-TDG-BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26932196',
'doi' => '',
'modified' => '2016-09-23 16:34:57',
'created' => '2016-09-23 16:34:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 71 => array(
'id' => '2833',
'name' => 'Genome-wide DNA methylation profile of developing deciduous tooth germ in miniature pigs',
'authors' => 'Su Y, Fan Z, Wu X, Li Y, Wang F, Zhang C, Wang J, Du J, Wang S',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND"><span class="highlight">DNA</span> <span class="highlight">methylation</span> is an important epigenetic modification critical to the regulation of gene expression during development. To date, little is known about the role of <span class="highlight">DNA</span> <span class="highlight">methylation</span> in <span class="highlight">tooth</span> development in large animal models. Thus, we carried out a comparative genomic analysis of <span class="highlight">genome-wide</span> <span class="highlight">DNA</span> <span class="highlight">methylation</span> profiles in E50 and E60 <span class="highlight">tooth</span> <span class="highlight">germ</span> from <span class="highlight">miniature</span> <span class="highlight">pigs</span> using methylated <span class="highlight">DNA</span> immunoprecipitation-sequencing (MeDIP-seq).</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We observed different <span class="highlight">DNA</span> <span class="highlight">methylation</span> patterns during the different developmental stages of pig <span class="highlight">tooth</span> <span class="highlight">germ</span>. A total of 2469 differentially methylated genes were identified. Functional analysis identified several signaling pathways and 104 genes that may be potential key regulators of pig <span class="highlight">tooth</span> development from E50 to E60.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">The present study provided a comprehensive analysis of the global <span class="highlight">DNA</span> <span class="highlight">methylation</span> pattern of <span class="highlight">tooth</span> <span class="highlight">germ</span> in <span class="highlight">miniature</span> <span class="highlight">pigs</span> and identified candidate genes that potentially regulate <span class="highlight">tooth</span> development from E50 to E60.</abstracttext></p>
</div>',
'date' => '2016-02-24',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26911717',
'doi' => '10.1186/s12864-016-2485-9',
'modified' => '2016-03-01 10:35:04',
'created' => '2016-03-01 10:35:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 72 => array(
'id' => '2834',
'name' => 'Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.',
'authors' => 'Rebollo R, Mager DL',
'description' => '<p>Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method used in our laboratory to detect, quantify, and compare mammalian endogenous retrovirus DNA methylation. More specifically we describe methylated DNA immunoprecipitation (MeDIP) followed by quantitative PCR.</p>',
'date' => '2016-02-20',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26895065',
'doi' => '10.1007/978-1-4939-3372-3_23',
'modified' => '2016-03-02 10:37:15',
'created' => '2016-03-02 10:22:16',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 73 => array(
'id' => '2815',
'name' => 'The dual specificity phosphatase 2 gene is hypermethylated in human cancer and regulated by epigenetic mechanisms',
'authors' => 'Tanja Haag, Antje M. Richter, Martin B. Schneider, Adriana P. Jiménez and Reinhard H. Dammann',
'description' => '<p><span>Dual specificity phosphatases are a class of tumor-associated proteins involved in the negative regulation of the MAP kinase pathway. Downregulation of the </span><em xmlns="" class="EmphasisTypeItalic">dual specificity phosphatase 2</em><span> (</span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span>) has been reported in cancer. Epigenetic silencing of tumor suppressor genes by abnormal promoter methylation is a frequent mechanism in oncogenesis. It has been shown that the epigenetic factor CTCF is involved in the regulation of tumor suppressor genes.</span></p>
<p><span>We analyzed the promoter hypermethylation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> in human cancer, including primary Merkel cell carcinoma by bisulfite restriction analysis and pyrosequencing. Moreover we analyzed the impact of a DNA methyltransferase inhibitor (5-Aza-dC) and CTCF on the epigenetic regulation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> by qRT-PCR, promoter assay, chromatin immuno-precipitation and methylation analysis.</span></span></p>
<p><span><span>Here we report a significant tumor-specific hypermethylation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> in primary Merkel cell carcinoma (</span><em xmlns="" class="EmphasisTypeItalic">p</em><span> = 0.05). An increase in methylation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> was also found in 17 out of 24 (71 %) cancer cell lines, including skin and lung cancer. Treatment of cancer cells with 5-Aza-dC induced </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression by its promoter demethylation, Additionally we observed that CTCF induces </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression in cell lines that exhibit silencing of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span>. This reactivation was accompanied by increased CTCF binding and demethylation of the </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> promoter.</span></span></span></p>
<p><span><span><span>Our data show that aberrant epigenetic inactivation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> occurs in carcinogenesis and that CTCF is involved in the epigenetic regulation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression.</span></span></span></span></p>',
'date' => '2016-02-01',
'pmid' => 'http://pubmed.gov/26833217',
'doi' => '10.1186/s12885-016-2087-6',
'modified' => '2016-03-09 16:34:51',
'created' => '2016-02-09 14:47:36',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 74 => array(
'id' => '2859',
'name' => 'Role of Growth Arrest and DNA Damage-Inducible, Beta in Alcohol-Drinking Behaviors',
'authors' => 'Gavin DP, Kusumo H, Zhang H, Guidotti A, Pandey SC',
'description' => '<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">The contribution of epigenetic factors, such as histone acetylation and DNA methylation, to the regulation of alcohol-drinking behavior has been increasingly recognized over the last several years. GADD45b is a protein demonstrated to be involved in DNA demethylation at neurotrophic factor gene promoters, including at brain-derived neurotrophic factor (Bdnf) which has been highly implicated in alcohol-drinking behavior.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">DNA methyltransferase-1 (Dnmt1), 3a, and 3b, and Gadd45a, b, and g mRNA were measured in the nucleus accumbens (NAc) and ventral tegmental areas of high ethanol (EtOH) consuming C57BL/6J (C57) and low alcohol consuming DBA/2J (DBA) mice using quantitative reverse transcriptase polymerase chain reaction (PCR). In the NAc, GADD45b protein was measured via immunohistochemistry and Bdnf9a mRNA using in situ PCR. Bdnf9a promoter histone H3 acetylated at lysines 9 and 14 (H3K9,K14ac) was measured using chromatin immunoprecipitation, and 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) using methylated DNA immunoprecipitation. Alcohol-drinking behavior was evaluated in Gadd45b haplodeficient (+/-) and null mice (-/-) utilizing drinking-in-the-dark (DID) and 2-bottle free-choice paradigms.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">C57 mice had lower levels of Gadd45b and g mRNA and GADD45b protein in the NAc relative to the DBA strain. C57 mice had lower NAc shell Bdnf9a mRNA levels, Bdnf9a promoter H3K9,K14ac, and higher Bdnf9a promoter 5HMC and 5MC. Acute EtOH increased GADD45b protein, Bdnf9a mRNA, and histone acetylation and decreased 5HMC in C57 mice. Gadd45b +/- mice displayed higher drinking behavior relative to wild-type littermates in both DID and 2-bottle free-choice paradigms.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">These data indicate the importance of the DNA demethylation pathway and its interactions with histone posttranslational modifications in alcohol-drinking behavior. Further, we suggest that lower DNA demethylation protein GADD45b levels may affect Bdnf expression possibly leading to altered alcohol-drinking behavior.</abstracttext></p>',
'date' => '2016-02-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26842245',
'doi' => ' 10.1111/acer.12965',
'modified' => '2016-03-15 16:37:22',
'created' => '2016-03-15 16:37:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 75 => array(
'id' => '2975',
'name' => 'Protocol for Methylated DNA Immunoprecipitation (MeDIP) Analysis',
'authors' => 'Karpova NN et al.',
'description' => '<p>DNA methylation is a fundamental epigenetic mechanism for silencing gene expression by either modifying chromatin structure to a repressive state or interfering with the transcription factors’ binding. DNA methylation primarily occurs at the position C5 of a cytosine ring mainly in the context of CpG dinucleotides. The modification can be recognized both in vivo and in vitro by the methyl-CpG binding proteins (MBPs) as well as in vitro by an antibody raised against 5-methylcytosine (5mC). This chapter describes different MBPs and introduces a standard methylated DNA immunoprecipitation (MeDIP) method, which is based on using the anti-5mC antibody to isolate methylated DNA fragments for subsequent locus-specific DNA methylation analysis. The MeDIP-generated DNA can be used as well for methylation profiling on a genome scale using array-based (MeDIP-chip) and high-throughput (MeDIP-seq) technologies.</p>',
'date' => '2016-02-01',
'pmid' => 'http://link.springer.com/protocol/10.1007/978-1-4939-2754-8_6',
'doi' => '10.1007/978-1-4939-2754-8_6',
'modified' => '2016-07-07 09:35:44',
'created' => '2016-07-07 09:35:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 76 => array(
'id' => '2844',
'name' => 'De novo DNA methylation drives 5hmC accumulation in mouse zygotes',
'authors' => 'Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, Koseki H, Sasaki H, Hajkova P',
'description' => '<p>Zygotic epigenetic reprogramming entails genome-wide DNA demethylation that is accompanied by Tet methylcytosine dioxygenase 3 (Tet3)-driven oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC; refs ,,,). Here we demonstrate using detailed immunofluorescence analysis and ultrasensitive LC-MS-based quantitative measurements that the initial loss of paternal 5mC does not require 5hmC formation. Small-molecule inhibition of Tet3 activity, as well as genetic ablation, impedes 5hmC accumulation in zygotes without affecting the early loss of paternal 5mC. Instead, 5hmC accumulation is dependent on the activity of zygotic Dnmt3a and Dnmt1, documenting a role for Tet3-driven hydroxylation in targeting de novo methylation activities present in the early embryo. Our data thus provide further insights into the dynamics of zygotic reprogramming, revealing an intricate interplay between DNA demethylation, de novo methylation and Tet3-driven hydroxylation.</p>',
'date' => '2016-01-11',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26751286',
'doi' => '10.1038/ncb3296',
'modified' => '2016-03-09 17:32:33',
'created' => '2016-03-09 17:32:33',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 77 => array(
'id' => '2860',
'name' => 'DNA methylation profiling: comparison of genome-wide sequencing methods and the Infinium Human Methylation 450 Bead Chip',
'authors' => 'Walker DL, Bhagwate AV, Baheti S, Smalley RL, Hilker CA, Sun Z, Cunningham JM',
'description' => '<div class="">
<h4>AIMS:</h4>
<p><abstracttext label="AIMS" nlmcategory="OBJECTIVE">To compare the performance of four sequence-based and one microarray methods for DNA methylation profiling.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">DNA from two cell lines were profiled by reduced representation bisulfite sequencing, methyl capture sequencing (SS-Meth Seq), NimbleGen SeqCapEpi CpGiant(Nimblegen MethSeq), methylated DNA immunoprecipitation (MeDIP) and the Human Methylation 450 Bead Chip (Meth450K).</abstracttext></p>
<h4>RESULTS & CONCLUSION:</h4>
<p><abstracttext label="RESULTS & CONCLUSION" nlmcategory="CONCLUSIONS">Despite differences in genome-wide coverage, high correlation and concordance were observed between different methods. Significant overlap of differentially methylated regions was identified between sequenced-based platforms. MeDIP provided the best coverage for the whole genome and gene body regions, while RRBS and Nimblegen MethSeq were superior for CpGs in CpG islands and promoters. Methylation analyses can be achieved by any of the five methods but understanding their differences may better address the research question being posed.</abstracttext></p>
</div>',
'date' => '2015-12-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26192535',
'doi' => '10.2217/EPI.15.64',
'modified' => '2016-03-16 11:06:05',
'created' => '2016-03-16 11:06:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 78 => array(
'id' => '2931',
'name' => 'Oxidative DNA damage in mouse sperm chromosomes: Size matters.',
'authors' => 'Kocer A et al.',
'description' => '<p>Normal embryo and foetal development as well as the health of the progeny are mostly dependent on gamete nuclear integrity. In the present study, in order to characterize more precisely oxidative DNA damage in mouse sperm we used two mouse models that display high levels of sperm oxidative DNA damage, a common alteration encountered both in in vivo and in vitro reproduction. Immunoprecipitation of oxidized sperm DNA coupled to deep sequencing showed that mouse chromosomes may be largely affected by oxidative alterations. We show that the vulnerability of chromosomes to oxidative attack inversely correlated with their size and was not linked to their GC richness. It was neither correlated with the chromosome content in persisting nucleosomes nor associated with methylated sequences. A strong correlation was found between oxidized sequences and sequences rich in short interspersed repeat elements (SINEs). Chromosome position in the sperm nucleus as revealed by fluorescent in situ hybridization appears to be a confounder. These data map for the first time fragile mouse sperm chromosomal regions when facing oxidative damage that may challenge the repair mechanisms of the oocyte post-fertilization.</p>',
'date' => '2015-12-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26510519',
'doi' => '10.1016/j.freeradbiomed.2015.10.419',
'modified' => '2016-05-19 10:18:24',
'created' => '2016-05-19 10:18:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 79 => array(
'id' => '2944',
'name' => 'Immunohistochemical Detection of Oxidized Forms of 5-Methylcytosine in Embryonic and Adult Brain Tissue',
'authors' => 'Abakir A et al.',
'description' => '<p>DNA methylation (5-methylcytosine, 5mC) is a major epigenetic modification of the eukaryotic genome associated with gene repression. Ten-eleven translocation proteins (Tet1/2/3) can oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Recent studies demonstrate that 5hmC is particularly enriched in neuronal cells and imply the involvement of this mark in transcriptional regulation taking place within the mammalian brain. Although a number of biochemical and antibody-based approaches have been successfully used to study the global content and genomic distributions of 5hmC in various contexts, most of these techniques do not provide any spatial information on the levels of this mark in different cell types. Here we describe a method of sensitive immunochemical detection of 5hmC/5fC/5caC in brain tissue based on the use of peroxidase-conjugated secondary antibodies and tyramide signal amplification. This technique can be instrumental in determining the relative enrichments of oxidized forms of 5mC in different brain cell types, effectively complementing other established approaches to investigate the functions of these marks in embryonic and adult brain.</p>',
'date' => '2015-09-02',
'pmid' => 'http://link.springer.com/protocol/10.1007%2F978-1-4939-2754-8_8',
'doi' => ' Print ISBN 978-1-4939-2753-1 Online ISBN 978-1-4939-2754-8 Series Title Neuromethods Series Volume 105 Series ISSN 0893-2336 Publisher Springer New York Copyright Holder Springer Science+Business Media New York Additional Lin',
'modified' => '2016-06-08 10:16:17',
'created' => '2016-06-08 10:16:17',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 80 => array(
'id' => '2847',
'name' => 'Gadd45b and N-methyl-D-aspartate induced DNA demethylation in postmitotic neurons.',
'authors' => 'Gavin DP, Kusumo H, Sharma RP, Guizzetti M, Guidotti A, Pandey SC.',
'description' => '<p><strong>AIM:</strong> In nondividing neurons examine the role of Gadd45b in active 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) removal at a gene promoter highly implicated in mental illnesses and cognition, Bdnf.</p>
<p><strong>MATERIALS & METHODS:</strong> Mouse primary cortical neuronal cultures with and without Gadd45b siRNA transfection were treated with N-methyl-d-aspartate (NMDA). Expression changes of genes reportedly involved in DNA demethylation, Bdnf mRNA and protein and 5MC and 5HMC at Bdnf promoters were measured.</p>
<p><strong>RESULTS:</strong> Gadd45b siRNA transfection in neurons abolishes the NMDA-induced increase in Bdnf IXa mRNA and reductions in 5MC and 5HMC at the Bdnf IXa promoter.</p>
<p><strong>CONCLUSION:</strong> These results contribute to our understanding of DNA demethylation mechanisms in neurons, and its role in regulating NMDA responsive genes implicated in mental illnesses.</p>',
'date' => '2015-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/26111030',
'doi' => '10.2217/epi.15.12',
'modified' => '2016-03-11 16:02:08',
'created' => '2016-03-11 15:47:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 81 => array(
'id' => '2879',
'name' => 'Active human nucleolar organizer regions are interspersed with inactive rDNA repeats in normal and tumor cells.',
'authors' => 'Zillner K, Komatsu J, Filarsky K, Kalepu R, Bensimon A, Németh A',
'description' => '<div class="">
<h4>AIM:</h4>
<p><abstracttext label="AIM" nlmcategory="OBJECTIVE">The synthesis of rRNA is a key determinant of normal and malignant cell growth and subject to epigenetic regulation. Yet, the epigenomic features of rDNA arrays clustered in nucleolar organizer regions are largely unknown. We set out to explore for the first time how DNA methylation is distributed on individual rDNA arrays.</abstracttext></p>
<h4>MATERIALS & METHODS:</h4>
<p><abstracttext label="MATERIALS & METHODS" nlmcategory="METHODS">Here we combined immunofluorescence detection of DNA modifications with fluorescence hybridization of single DNA fibers, metaphase immuno-FISH and methylation-sensitive restriction enzyme digestions followed by Southern blot.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We found clustering of both hypomethylated and hypermethylated repeat units and hypermethylation of noncanonical rDNA in IMR90 fibroblasts and HCT116 colorectal carcinoma cells. Surprisingly, we also found transitions between hypo- and hypermethylated rDNA repeat clusters on single DNA fibers.</abstracttext></p>
<h4>CONCLUSION:</h4>
<p><abstracttext label="CONCLUSION" nlmcategory="CONCLUSIONS">Collectively, our analyses revealed co-existence of different epialleles on individual nucleolar organizer regions and showed that epi-combing is a valuable approach to analyze epigenomic patterns of repetitive DNA.</abstracttext></p>
</div>',
'date' => '2015-06-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26077426',
'doi' => '10.2217/epi.14.93',
'modified' => '2016-04-05 09:44:29',
'created' => '2016-04-05 09:44:29',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 82 => array(
'id' => '2790',
'name' => 'Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency.',
'authors' => 'Chen H, Aksoy I, Gonnot F, Osteil P, Aubry M, Hamela C, Rognard C, Hochard A, Voisin S, Fontaine E, Mure M, Afanassieff M, Cleroux E, Guibert S, Chen J, Vallot C, Acloque H, Genthon C, Donnadieu C, De Vos J, Sanlaville D, Guérin JF, Weber M, Stanton LW, R',
'description' => 'Leukemia inhibitory factor (LIF)/STAT3 signalling is a hallmark of naive pluripotency in rodent pluripotent stem cells (PSCs), whereas fibroblast growth factor (FGF)-2 and activin/nodal signalling is required to sustain self-renewal of human PSCs in a condition referred to as the primed state. It is unknown why LIF/STAT3 signalling alone fails to sustain pluripotency in human PSCs. Here we show that the forced expression of the hormone-dependent STAT3-ER (ER, ligand-binding domain of the human oestrogen receptor) in combination with 2i/LIF and tamoxifen allows human PSCs to escape from the primed state and enter a state characterized by the activation of STAT3 target genes and long-term self-renewal in FGF2- and feeder-free conditions. These cells acquire growth properties, a gene expression profile and an epigenetic landscape closer to those described in mouse naive PSCs. Together, these results show that temporarily increasing STAT3 activity is sufficient to reprogramme human PSCs to naive-like pluripotent cells.',
'date' => '2015-05-13',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25968054',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 83 => array(
'id' => '2678',
'name' => 'Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells.',
'authors' => 'Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, Pop R, Reyon D, Tsai SQ, Mallard W, Joung JK, Rinn JL, Gnirke A, Meissner A',
'description' => 'DNA methylation is a key epigenetic modification involved in regulating gene expression and maintaining genomic integrity. Here we inactivated all three catalytically active DNA methyltransferases (DNMTs) in human embryonic stem cells (ESCs) using CRISPR/Cas9 genome editing to further investigate the roles and genomic targets of these enzymes. Disruption of DNMT3A or DNMT3B individually as well as of both enzymes in tandem results in viable, pluripotent cell lines with distinct effects on the DNA methylation landscape, as assessed by whole-genome bisulfite sequencing. Surprisingly, in contrast to findings in mouse, deletion of DNMT1 resulted in rapid cell death in human ESCs. To overcome this immediate lethality, we generated a doxycycline-responsive tTA-DNMT1* rescue line and readily obtained homozygous DNMT1-mutant lines. However, doxycycline-mediated repression of exogenous DNMT1* initiates rapid, global loss of DNA methylation, followed by extensive cell death. Our data provide a comprehensive characterization of DNMT-mutant ESCs, including single-base genome-wide maps of the targets of these enzymes.',
'date' => '2015-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25822089',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 84 => array(
'id' => '2477',
'name' => 'Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes.',
'authors' => 'Li L, Zhang Y, Fan Y, Sun K, Su X, Du Z, Tsao SW, Loh TK, Sun H, Chan AT, Zeng YX, Chan WY, Chan FK, Tao Q',
'description' => 'Aims: Nasopharyngeal carcinoma (NPC) is a common tumor consistently associated with Epstein-Barr virus infection and prevalent in South China, including Hong Kong, and southeast Asia. Current genomic sequencing studies found only rare mutations in NPC, indicating its critical epigenetic etiology, while no epigenome exists for NPC as yet. Materials & methods: We profiled the methylomes of NPC cell lines and primary tumors, together with normal nasopharyngeal epithelial cells, using methylated DNA immunoprecipitation (MeDIP). Results: We observed extensive, genome-wide methylation of cellular genes. Epigenetic disruption of Wnt, MAPK, TGF-β and Hedgehog signaling pathways was detected. Methylation of Wnt signaling regulators (SFRP1, 2, 4 and 5, DACT2, DKK2 and DKK3) was frequently detected in tumor and nasal swab samples from NPC patients. Functional studies showed that these genes are bona fide tumor-suppressor genes for NPC. Conclusion: The NPC methylome shows a special high-degree CpG methylation epigenotype, similar to the Epstein-Barr virus-infected gastric cancer, indicating a critical epigenetic etiology for NPC pathogenesis.',
'date' => '2015-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25479246',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 85 => array(
'id' => '2613',
'name' => 'CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.',
'authors' => 'Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC',
'description' => 'BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the underlying epigenetic signatures involved in its regulation. RESULTS: NOS3 and STAT3 mRNA levels were elevated in HUAEC of patients who suffered from placental insufficiency. 5-hydroxymethylcytosine, H3K9ac and Histone 2A (H2A).Zac at the NOS3 transcription start site directly correlated with NOS3 mRNA levels. Concomitantly, we observed entangled histone acetylation patterns and NOS3 response upon hypoxic conditions in vitro. Knock-down of either NOS3 or STAT3 by RNAi provided evidence for a functional NOS3/STAT3 relationship. Moreover, we recognized massive turnover of Stat3 at a discrete binding site in the NOS3 promoter. Interestingly, induced hyperacetylation resulted in short-termed increase of NOS3 mRNA followed by deferred decrease indicating that NOS3 expression could become self-attenuated by co-expressed intronic 27 nt-ncRNA. Reporter assay results and phylogenetic analyses enabled us to propose a novel model for STAT3-3'-UTR targeting by this 27-nt-ncRNA. CONCLUSIONS: An adverse intrauterine environment leads to adaptive changes of NOS3 expression. Apparently, a rapid NOS3 self-limiting response upon ectopic triggers co-exists with longer termed expression changes in response to placental insufficiency involving differential epigenetic signatures. Their persistence might contribute to impaired vascular endothelial response and consequently increase the risk of cardiovascular disease later in life.',
'date' => '2015-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25699114',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 86 => array(
'id' => '2437',
'name' => 'Acute Depletion Redefines the Division of Labor among DNA Methyltransferases in Methylating the Human Genome.',
'authors' => 'Tiedemann RL, Putiri EL, Lee JH, Hlady RA, Kashiwagi K, Ordog T, Zhang Z, Liu C, Choi JH, Robertson KD',
'description' => 'Global patterns of DNA methylation, mediated by the DNA methyltransferases (DNMTs), are disrupted in all cancers by mechanisms that remain largely unknown, hampering their development as therapeutic targets. Combinatorial acute depletion of all DNMTs in a pluripotent human tumor cell line, followed by epigenome and transcriptome analysis, revealed DNMT functions in fine detail. DNMT3B occupancy regulates methylation during differentiation, whereas an unexpected interplay was discovered in which DNMT1 and DNMT3B antithetically regulate methylation and hydroxymethylation in gene bodies, a finding confirmed in other cell types. DNMT3B mediated non-CpG methylation, whereas DNMT3L influenced the activity of DNMT3B toward non-CpG versus CpG site methylation. Altogether, these data reveal functional targets of each DNMT, suggesting that isoform selective inhibition would be therapeutically advantageous.',
'date' => '2014-11-20',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25453758',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 87 => array(
'id' => '2368',
'name' => 'A B-cell targeting virus disrupts potentially protective genomic methylation patterns in lymphoid tissue by increasing global 5-hydroxmethylcytosine levels',
'authors' => 'Ciccone NA, Mwangi W, Ruzov A, Smith LP, Butter C, Nair V',
'description' => 'The mechanisms by which viruses modulate the immune system include changes in host genomic methylation. 5-hydroxmethylcytosine (5hmC) is the catalytic product of the Tet (Ten-11 translocation) family of enzymes and may serve as an intermediate of DNA demethylation. Recent reports suggest that 5hmC may confer consequences on cellular events including the pathogenesis of disease; in order to explore this possibility further we investigated both 5-methylcytosine (5mC) and 5hmC levels in healthy and diseased chicken bursas of Fabricius. We discovered that embryonic B-cells have high 5mC content while 5hmC decreases during bursa development. We propose that a high 5mC level protects from the mutagenic activity of the B-cell antibody diversifying enzyme activation induced deaminase (AID). In support of this view, AID mRNA increases significantly within the developing bursa from embryonic to post hatch stages while mRNAs that encode Tet family members 1 and 2 reduce over the same period. Moreover, our data revealed that infectious bursal disease virus (IBDV) disrupts this genomic methylation pattern causing a global increase in 5hmC levels in a mechanism that may involve increased Tet 1 and 2 mRNAs. To our knowledge this is the first time that a viral infection has been observed to cause global increases in genomic 5hmC within infected host tissues, underlining a mechanism that may involve the induction of B-cell genomic instability and cell death to facilitate viral egress.',
'date' => '2014-10-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/25338704',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 88 => array(
'id' => '2350',
'name' => 'Spontaneous sleep-wake cycle and sleep deprivation differently induce Bdnf1, Bdnf4 and Bdnf9a DNA methylation and transcripts levels in the basal forebrain and frontal cortex in rats.',
'authors' => 'Ventskovska O, Porkka-Heiskanen T, Karpova NN',
'description' => 'Brain-derived neurotrophic factor (Bdnf) regulates neuronal plasticity, slow wave activity and sleep homeostasis. Environmental stimuli control Bdnf expression through epigenetic mechanisms, but there are no data on epigenetic regulation of Bdnf by sleep or sleep deprivation. Here we investigated whether 5-methylcytosine (5mC) DNA modification at Bdnf promoters p1, p4 and p9 influences Bdnf1, Bdnf4 and Bdnf9a expression during the normal inactive phase or after sleep deprivation (SD) (3, 6 and 12 h, end-times being ZT3, ZT6 and ZT12) in rats in two brain areas involved in sleep regulation, the basal forebrain and cortex. We found a daytime variation in cortical Bdnf expression: Bdnf1 expression was highest at ZT6 and Bdnf4 lowest at ZT12. Such variation was not observed in the basal forebrain. Also Bdnf p1 and p9 methylation levels differed only in the cortex, while Bdnf p4 methylation did not vary in either area. Factorial analysis revealed that sleep deprivation significantly induced Bdnf1 and Bdnf4 with the similar pattern for Bdnf9a in both basal forebrain and cortex; 12 h of sleep deprivation decreased 5mC levels at the cortical Bdnf p4 and p9. Regression analysis between the 5mC promoter levels and the corresponding Bdnf transcript expression revealed significant negative correlations for the basal forebrain Bdnf1 and cortical Bdnf9a transcripts in only non-deprived rats, while these correlations were lost after sleep deprivation. Our results suggest that Bdnf transcription during the light phase of undisturbed sleep-wake cycle but not after SD is regulated at least partially by brain site-specific DNA methylation.',
'date' => '2014-09-15',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25223586',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 89 => array(
'id' => '2109',
'name' => 'Transient accumulation of 5-carboxylcytosine indicates involvement of active demethylation in lineage specification of neural stem cells.',
'authors' => 'Wheldon LM, Abakir A, Ferjentsik Z, Dudnakova T, Strohbuecker S, Christie D, Dai N, Guan S, Foster JM, Corrêa IR, Loose M, Dixon JE, Sottile V, Johnson AD, Ruzov A',
'description' => '5-Methylcytosine (5mC) is an epigenetic modification involved in regulation of gene activity during differentiation. Tet dioxygenases oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). Both 5fC and 5caC can be excised from DNA by thymine-DNA glycosylase (TDG) followed by regeneration of unmodified cytosine via the base excision repair pathway. Despite evidence that this mechanism is operative in embryonic stem cells, the role of TDG-dependent demethylation in differentiation and development is currently unclear. Here, we demonstrate that widespread oxidation of 5hmC to 5caC occurs in postimplantation mouse embryos. We show that 5fC and 5caC are transiently accumulated during lineage specification of neural stem cells (NSCs) in culture and in vivo. Moreover, 5caC is enriched at the cell-type-specific promoters during differentiation of NSCs, and TDG knockdown leads to increased 5fC/5caC levels in differentiating NSCs. Our data suggest that active demethylation contributes to epigenetic reprogramming determining lineage specification in embryonic brain.',
'date' => '2014-06-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24882006',
'doi' => '',
'modified' => '2015-07-24 15:39:03',
'created' => '2015-07-24 15:39:03',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 90 => array(
'id' => '2448',
'name' => 'Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring.',
'authors' => 'Itzhak Y, Ergui I, Young JI',
'description' => 'The high rate of methamphetamine (METH) abuse among young adults and women of childbearing age makes it imperative to determine the long-term effects of METH exposure on the offspring. We hypothesized that parental METH exposure modulates offspring behavior by disrupting epigenetic programming of gene expression in the brain. To simulate the human pattern of drug use, male and female C57Bl/6J mice were exposed to escalating doses of METH or saline from adolescence through adulthood; following mating, females continue to receive drug or saline through gestational day 17. F1 METH male offspring showed enhanced response to cocaine-conditioned reward and hyperlocomotion. Both F1 METH male and female offspring had reduced response to conditioned fear. Cross-fostering experiments have shown that certain behavioral phenotypes were modulated by maternal care of either METH or saline dams. Analysis of offspring hippocampal DNA methylation showed differentially methylated regions as a result of both METH in utero exposure and maternal care. Our results suggest that behavioral phenotypes and epigenotypes of offspring that were exposed to METH in utero are vulnerable to (a) METH exposure during embryonic development, a period when wide epigenetic reprogramming occurs, and (b) postnatal maternal care.Molecular Psychiatry advance online publication, 18 February 2014; doi:10.1038/mp.2014.7.',
'date' => '2014-02-18',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24535458',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 91 => array(
'id' => '1843',
'name' => 'Alterations of epigenetic signatures in hepatocyte nuclear factor 4α deficient mouse liver determined by improved ChIP-qPCR and (h)MeDIP-qPCR assays.',
'authors' => 'Zhang Q, Lei X, Lu H',
'description' => 'Hepatocyte nuclear factor 4α (HNF4α) is a liver-enriched transcription factor essential for liver development and function. In hepatocytes, HNF4α regulates a large number of genes important for nutrient/xenobiotic metabolism and cell differentiation and proliferation. Currently, little is known about the epigenetic mechanism of gene regulation by HNF4α. In this study, the global and specific alterations at the selected gene loci of representative histone modifications and DNA methylations were investigated in Hnf4a-deficient female mouse livers using the improved MeDIP-, hMeDIP- and ChIP-qPCR assay. Hnf4a deficiency significantly increased hepatic total IPed DNA fragments for histone H3 lysine-4 dimethylation (H3K4me2), H3K4me3, H3K9me2, H3K27me3 and H3K4 acetylation, but not for H3K9me3, 5-methylcytosine,or 5-hydroxymethylcytosine. At specific gene loci, the relative enrichments of histone and DNA modifications were changed to different degree in Hnf4a-deficient mouse liver. Among the epigenetic signatures investigated, changes in H3K4me3 correlated the best with mRNA expression. Additionally, Hnf4a-deficient livers had increased mRNA expression of histone H1.2 and H3.3 as well as epigenetic modifiers Dnmt1, Tet3, Setd7, Kmt2c, Ehmt2, and Ezh2. In conclusion, the present study provides convenient improved (h)MeDIP- and ChIP-qPCR assays for epigenetic study. Hnf4a deficiency in young-adult mouse liver markedly alters histone methylation and acetylation, with fewer effects on DNA methylation and 5-hydroxymethylation. The underlying mechanism may be the induction of epigenetic enzymes responsible for the addition/removal of the epigenetic signatures, and/or the loss of HNF4α per se as a key coordinator for epigenetic modifiers.',
'date' => '2014-01-10',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24427299',
'doi' => '',
'modified' => '2015-07-24 15:39:01',
'created' => '2015-07-24 15:39:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 92 => array(
'id' => '1773',
'name' => 'Peroxisome proliferator-activated receptor γ regulates genes involved in insulin/insulin-like growth factor signaling and lipid metabolism during adipogenesis through functionally distinct enhancer classes.',
'authors' => 'Oger F, Dubois-Chevalier J, Gheeraert C, Avner S, Durand E, Froguel P, Salbert G, Staels B, Lefebvre P, Eeckhoute J',
'description' => 'The nuclear receptor peroxisome proliferator-activated receptor (PPAR) is a transcription factor whose expression is induced during adipogenesis and that is required for the acquisition and control of mature adipocyte functions. Indeed, PPAR induces the expression of genes involved in lipid synthesis and storage through enhancers activated during adipocyte differentiation. Here, we show that PPAR also binds to enhancers already active in preadipocytes as evidenced by an active chromatin state including lower DNA methylation levels despite higher CpG content. These constitutive enhancers are linked to genes involved in the insulin/insulin-like growth factor signaling pathway that are transcriptionally induced during adipogenesis but to a lower extent than lipid metabolism genes, because of stronger basal expression levels in preadipocytes. This is consistent with the sequential involvement of hormonal sensitivity and lipid handling during adipocyte maturation and correlates with the chromatin structure dynamics at constitutive and activated enhancers. Interestingly, constitutive enhancers are evolutionary conserved and can be activated in other tissues, in contrast to enhancers controlling lipid handling genes whose activation is more restricted to adipocytes. Thus, PPAR utilizes both broadly active and cell type-specific enhancers to modulate the dynamic range of activation of genes involved in the adipogenic process.',
'date' => '2014-01-10',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24288131',
'doi' => '',
'modified' => '2015-07-24 15:39:01',
'created' => '2015-07-24 15:39:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 93 => array(
'id' => '1572',
'name' => 'Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model.',
'authors' => 'Yafune A, Kawai M, Itahashi M, Kimura M, Nakane F, Mitsumori K, Shibutani M',
'description' => 'Disruptive epigenetic gene control has been shown to be involved in carcinogenesis. To identify key molecules in piperonyl butoxide (PBO)-induced hepatocarcinogenesis, we searched hypermethylated genes using CpG island (CGI) microarrays in non-neoplastic liver cells as a source of proliferative lesions at 25 weeks after tumor promotion with PBO using mice. We further performed methylation-specific polymerase chain reaction (PCR), real-time reverse transcription PCR, and immunohistochemical analysis in PBO-promoted liver tissues. Ebp4.1, Wdr6 and Cmtm6 increased methylation levels in the promoter region by PBO promotion, although Cmtm6 levels were statistically non-significant. These results suggest that PBO promotion may cause altered epigenetic gene regulation in non-neoplastic liver cells surrounding proliferative lesions to allow the facilitation of hepatocarcinogenesis. Both Wdr6 and Cmtm6 showed decreased expression in non-neoplastic liver cells in contrast to positive immunoreactivity in the majority of proliferative lesions produced by PBO promotion. These results suggest that both Wdr6 and Cmtm6 were spared from epigenetic gene modification in proliferative lesions by PBO promotion in contrast to the hypermethylation-mediated downregulation in surrounding liver cells. Considering the effective detection of proliferative lesions, these molecules could be used as detection markers of hepatocellular proliferative lesions and played an important role in hepatocarcinogenesis.',
'date' => '2013-10-09',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23968726',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 94 => array(
'id' => '1569',
'name' => 'Genome-wide screening identifies Plasmodium chabaudi-induced modifications of DNA methylation status of Tlr1 and Tlr6 gene promoters in liver, but not spleen, of female C57BL/6 mice.',
'authors' => 'Al-Quraishy S, Dkhil MA, Abdel-Baki AA, Delic D, Santourlidis S, Wunderlich F',
'description' => '<p>Epigenetic reprogramming of host genes via DNA methylation is increasingly recognized as critical for the outcome of diverse infectious diseases, but information for malaria is not yet available. Here, we investigate the effect of blood-stage malaria of Plasmodium chabaudi on the DNA methylation status of host gene promoters on a genome-wide scale using methylated DNA immunoprecipitation and Nimblegen microarrays containing 2,000 bp oligonucleotide features that were split into -1,500 to -500 bp Ups promoters and -500 to +500 bp Cor promoters, relative to the transcription site, for evaluation of differential DNA methylation. Gene expression was analyzed by Agilent and Affymetrix microarray technology. Challenging of female C57BL/6 mice with 10(6) P. chabaudi-infected erythrocytes resulted in a self-healing outcome of infections with peak parasitemia on day 8 p.i. These infections induced organ-specific modifications of DNA methylation of gene promoters. Among the 17,354 features on Nimblegen arrays, only seven gene promoters were identified to be hypermethylated in the spleen, whereas the liver exhibited 109 hyper- and 67 hypomethylated promoters at peak parasitemia in comparison with non-infected mice. Among the identified genes with differentially methylated Cor-promoters, only the 7 genes Pigr, Ncf1, Klkb1, Emr1, Ndufb11, and Tlr6 in the liver and Apol6 in the spleen were detected to have significantly changed their expression. Remarkably, the Cor promoter of the toll-like receptor Tlr6 became hypomethylated and Tlr6 expression increased by 3.4-fold during infection. Concomitantly, the Ups promoter of the Tlr1 was hypermethylated, but Tlr1 expression also increased by 11.3-fold. TLR6 and TLR1 are known as auxillary receptors to form heterodimers with TLR2 in plasma membranes of macrophages, which recognize different pathogen-associated molecular patterns (PAMPs), as, e.g., intact 3-acyl and sn-2-lyso-acyl glycosylphosphatidylinositols of P. falciparum, respectively. Our data suggest therefore that malaria-induced epigenetic fine-tuning of Tlr6 and Tlr1 through DNA methylation of their gene promoters in the liver is critically important for initial recognition of PAMPs and, thus, for the final self-healing outcome of blood-stage infections with P. chabaudi malaria.</p>',
'date' => '2013-08-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23949311',
'doi' => '',
'modified' => '2017-10-10 10:37:58',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 95 => array(
'id' => '1466',
'name' => 'Characterization of the DNA methylome and its interindividual variation in human peripheral blood monocytes.',
'authors' => 'Shen H, Qiu C, Li J, Tian Q, Deng HW',
'description' => 'AIM: Peripheral blood monocytes (PBMs) play multiple and critical roles in the immune response, and abnormalities in PBMs have been linked to a variety of human disorders. However, the DNA methylation landscape in PBMs is largely unknown. In this study, we characterized epigenome-wide DNA methylation profiles in purified PBMs. MATERIALS & METHODS: PBMs were isolated from freshly collected peripheral blood from 18 unrelated healthy postmenopausal Caucasian females. Epigenome-wide DNA methylation profiles (the methylome) were characterized by using methylated DNA immunoprecipitation combined with high-throughput sequencing. RESULTS: Distinct patterns were revealed at different genomic features. For instance, promoters were commonly (∼58%) found to be unmethylated; whereas protein coding regions were largely (∼84%) methylated. Although CpG-rich and -poor promoters showed distinct methylation patterns, interestingly, a negative correlation between promoter methylation levels and gene transcription levels was consistently observed across promoters with high to low CpG densities. Importantly, we observed substantial interindividual variation in DNA methylation across the individual PBM methylomes and the pattern of this interindividual variation varied between different genomic features, with highly variable regions enriched for repetitive DNA elements. Furthermore, we observed a modest but significant excess (p < 2.2 × 10(-16)) of genes showing a negative correlation between interindividual promoter methylation and transcription levels. These significant genes were enriched in biological processes that are closely related to PBM functions, suggesting that alteration in DNA methylation is likely to be an important mechanism contributing to the interindividual variation in PBM function, and PBM-related phenotypic and disease-susceptibility variation in humans. CONCLUSION: This study represents a comprehensive analysis of the human PBM methylome and its interindividual variation. Our data provide a valuable resource for future epigenomic and multiomic studies, exploring biological and disease-related regulatory mechanisms in PBMs.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23750642',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 96 => array(
'id' => '1463',
'name' => 'Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation.',
'authors' => 'Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD',
'description' => 'Histone post-translational modifications regulate chromatin structure and function largely through interactions with effector proteins that often contain multiple histone-binding domains. While significant progress has been made characterizing individual effector domains, the role of paired domains and how they function in a combinatorial fashion within chromatin are poorly defined. Here we show that the linked tandem Tudor and plant homeodomain (PHD) of UHRF1 (ubiquitin-like PHD and RING finger domain-containing protein 1) operates as a functional unit in cells, providing a defined combinatorial readout of a heterochromatin signature within a single histone H3 tail that is essential for UHRF1-directed epigenetic inheritance of DNA methylation. These findings provide critical support for the "histone code" hypothesis, demonstrating that multivalent histone engagement plays a key role in driving a fundamental downstream biological event in chromatin.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23752590',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 97 => array(
'id' => '1449',
'name' => 'Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs.',
'authors' => 'Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R',
'description' => 'The presence of 5-methylcytidine (m(5)C) in tRNA and rRNA molecules of a wide variety of organisms was first observed more than 40 years ago. However, detection of this modification was limited to specific, abundant, RNA species, due to the usage of low-throughput methods. To obtain a high resolution, systematic, and comprehensive transcriptome-wide overview of m(5)C across the three domains of life, we used bisulfite treatment on total RNA from both gram positive (B. subtilis) and gram negative (E. coli) bacteria, an archaeon (S. solfataricus) and a eukaryote (S. cerevisiae), followed by massively parallel sequencing. We were able to recover most previously documented m(5)C sites on rRNA in the four organisms, and identified several novel sites in yeast and archaeal rRNAs. Our analyses also allowed quantification of methylated m(5)C positions in 64 tRNAs in yeast and archaea, revealing stoichiometric differences between the methylation patterns of these organisms. Molecules of tRNAs in which m(5)C was absent were also discovered. Intriguingly, we detected m(5)C sites within archaeal mRNAs, and identified a consensus motif of AUCGANGU that directs methylation in S. solfataricus. Our results, which were validated using m(5)C-specific RNA immunoprecipitation, provide the first evidence for mRNA modifications in archaea, suggesting that this mode of post-transcriptional regulation extends beyond the eukaryotic domain.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23825970',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 98 => array(
'id' => '1403',
'name' => 'Methyl donor supplementation blocks the adverse effects of maternal high fat diet on offspring physiology.',
'authors' => 'Carlin J, George R, Reyes TM',
'description' => 'Maternal consumption of a high fat diet during pregnancy increases the offspring risk for obesity. Using a mouse model, we have previously shown that maternal consumption of a high fat (60%) diet leads to global and gene specific decreases in DNA methylation in the brain of the offspring. The present experiments were designed to attempt to reverse this DNA hypomethylation through supplementation of the maternal diet with methyl donors, and to determine whether methyl donor supplementation could block or attenuate phenotypes associated with maternal consumption of a HF diet. Metabolic and behavioral (fat preference) outcomes were assessed in male and female adult offspring. Expression of the mu-opioid receptor and dopamine transporter mRNA, as well as global DNA methylation were measured in the brain. Supplementation of the maternal diet with methyl donors attenuated the development of some of the adverse effects seen in offspring from dams fed a high fat diet; including weight gain, increased fat preference (males), changes in CNS gene expression and global hypomethylation in the prefrontal cortex. Notable sex differences were observed. These findings identify the importance of balanced methylation status during pregnancy, particularly in the context of a maternal high fat diet, for optimal offspring outcome.',
'date' => '2013-05-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23658839',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 99 => array(
'id' => '1311',
'name' => 'Naive pluripotency is associated with global DNA hypomethylation.',
'authors' => 'Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, Surani MA, Hajkova P',
'description' => 'Naive pluripotent embryonic stem cells (ESCs) and embryonic germ cells (EGCs) are derived from the preimplantation epiblast and primordial germ cells (PGCs), respectively. We investigated whether differences exist between ESCs and EGCs, in view of their distinct developmental origins. PGCs are programmed to undergo global DNA demethylation; however, we find that EGCs and ESCs exhibit equivalent global DNA methylation levels. Inhibition of MEK and Gsk3b by 2i conditions leads to pronounced reduction in DNA methylation in both cell types. This is driven by Prdm14 and is associated with downregulation of Dnmt3a and Dnmt3b. However, genomic imprints are maintained in 2i, and we report derivation of EGCs with intact genomic imprints. Collectively, our findings establish that culture in 2i instills a naive pluripotent state with a distinctive epigenetic configuration that parallels molecular features observed in both the preimplantation epiblast and nascent PGCs.',
'date' => '2013-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23416945',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 100 => array(
'id' => '1290',
'name' => 'DNA methylation analysis in the intestinal epithelium-effect of cell separation on gene expression and methylation profile.',
'authors' => 'Jenke AC, Postberg J, Raine T, Nayak KM, Molitor M, Wirth S, Kaser A, Parkes M, Heuschkel RB, Orth V, Zilbauer M',
'description' => 'BACKGROUND: Epigenetic signatures are highly cell type specific. Separation of distinct cell populations is therefore desirable for all epigenetic studies. However, to date little information is available on whether separation protocols might influence epigenetic and/or gene expression signatures and hence might be less beneficial. We investigated the influence of two frequently used protocols to isolate intestinal epithelium cells (IECs) from 6 healthy individuals. MATERIALS AND METHODS: Epithelial cells were isolated from small bowel (i.e. terminal ileum) biopsies using EDTA/DTT and enzymatic release followed by magnetic bead sorting via EPCAM labeled microbeads. Effects on gene/mRNA expression were analyzed using a real time PCR based expression array. DNA methylation was assessed by pyrosequencing of bisulfite converted DNA and methylated DNA immunoprecipitation (MeDIP). RESULTS: While cell purity was >95% using both cell separation approaches, gene expression analysis revealed significantly higher mRNA levels of several inflammatory genes in EDTA/DTT when compared to enzymatically released cells. In contrast, DNA methylation of selected genes was less variable and only revealed subtle differences. Comparison of DNA methylation of the epithelial cell marker EPCAM in unseparated whole biopsy samples with separated epithelium (i.e. EPCAM positive and negative fraction) demonstrated significant differences in DNA methylation between all three tissue fractions indicating cell type specific methylation patterns can be masked in unseparated tissue samples. CONCLUSIONS: Taken together, our data highlight the importance of considering the potential effect of cell separation on gene expression as well as DNA methylation signatures. The decision to separate tissue samples will therefore depend on study design and specific separation protocols.',
'date' => '2013-02-08',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23409010',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 101 => array(
'id' => '1065',
'name' => 'Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation.',
'authors' => 'Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD',
'description' => 'A fundamental challenge in mammalian biology has been the elucidation of mechanisms linking DNA methylation and histone post-translational modifications. Human UHRF1 (ubiquitin-like PHD and RING finger domain-containing 1) has multiple domains that bind chromatin, and it is implicated genetically in the maintenance of DNA methylation. However, molecular mechanisms underlying DNA methylation regulation by UHRF1 are poorly defined. Here we show that UHRF1 association with methylated histone H3 Lys9 (H3K9) is required for DNA methylation maintenance. We further show that UHRF1 association with H3K9 methylation is insensitive to adjacent H3 S10 phosphorylation-a known mitotic 'phospho-methyl switch'. Notably, we demonstrate that UHRF1 mitotic chromatin association is necessary for DNA methylation maintenance through regulation of the stability of DNA methyltransferase-1. Collectively, our results define a previously unknown link between H3K9 methylation and the faithful epigenetic inheritance of DNA methylation, establishing a notable mitotic role for UHRF1 in this process.',
'date' => '2012-09-30',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23022729',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 102 => array(
'id' => '960',
'name' => 'Histone acetylation and DNA demethylation of T-cells result in an anaplastic large cell lymphoma-like phenotype.',
'authors' => 'Joosten M, Seitz V, Zimmermann K, Sommerfeld A, Berg E, Lenze D, Leser U, Stein H, Hummel M',
'description' => 'Background. A characteristic feature of anaplastic large cell lymphoma is the significant repression of the T-cell expression program despite its T-cell origin. The reasons for this down-regulation of T-cell phenotype are still unknown. Design and Methods. To elucidate whether epigenetic mechanisms are responsible for the loss of the T-cell phenotype, we treated anaplastic large cell lymphoma and T-cell lymphoma/leukemia cell lines (n=4, each) with epigenetic modifiers to evoke DNA demethylation and histone acetylation. Global gene expression data from treated and untreated cell lines were generated and selected differentially expressed genes were evaluated by real-time RT-PCR and Western Blot analysis. Additionally, histone H3 lysine 27 trimethylation was analyzed by chromatin immunoprecipitation. Results. Combined DNA demethylation and histone acetylation of anaplastic large cell lymphoma cells was not able to reconstitute their T-cell phenotype. Instead, the same treatment induced in T-cells (i) an up-regulation of anaplastic large cell lymphoma-characteristic genes (e.g. ID2, LGALS1, c-JUN) and (ii) an almost complete extinction of their T-cell phenotype including CD3, LCK and ZAP70. In addition, a suppressive trimethylation of histone H3 lysine 27 of important T-cell transcription factor genes (GATA3, LEF1, TCF1) was present in anaplastic large cell lymphoma cells, which is in line with their absence in primary tumour specimens as demonstrated by immunohistochemistry. Conclusions. Our data suggest that epigenetically activated suppressors (e.g. ID2) contribute to the down-regulation of the T-cell expression program in anaplastic large cell lymphoma, which is maintained by trimethylation of histone H3 lysine 27.',
'date' => '2012-08-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22899583',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 103 => array(
'id' => '389',
'name' => 'Growth Arrest and DNA-Damage-Inducible, Beta (GADD45b)-Mediated DNA Demethylation in Major Psychosis.',
'authors' => 'Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A',
'description' => 'Aberrant neocortical DNA methylation has been suggested to be a pathophysiological contributor to psychotic disorders. Recently, a growth arrest and DNA-damage-inducible, beta (GADD45b) protein-coordinated DNA demethylation pathway, utilizing cytidine deaminases and thymidine glycosylases, has been identified in the brain. We measured expression of several members of this pathway in parietal cortical samples from the Stanley Foundation Neuropathology Consortium (SFNC) cohort. We find an increase in GADD45b mRNA and protein in patients with psychosis. In immunohistochemistry experiments using samples from the Harvard Brain Tissue Resource Center, we report an increased number of GADD45b-stained cells in prefrontal cortical layers II, III, and V in psychotic patients. Brain-derived neurotrophic factor IX (BDNF IXabcd) was selected as a readout gene to determine the effects of GADD45b expression and promoter binding. We find that there is less GADD45b binding to the BDNF IXabcd promoter in psychotic subjects. Further, there is reduced BDNF IXabcd mRNA expression, and an increase in 5-methylcytosine and 5-hydroxymethylcytosine at its promoter. On the basis of these results, we conclude that GADD45b may be increased in psychosis compensatory to its inability to access gene promoter regions.',
'date' => '2012-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22048458',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 104 => array(
'id' => '409',
'name' => 'Epigenetic silencing mediated through activated PI3K/AKT signaling in breast cancer.',
'authors' => 'Zuo T, Liu TM, Lan X, Weng YI, Shen R, Gu F, Huang YW, Liyanarachchi S, Deatherage DE, Hsu PY, Taslim C, Ramaswamy B, Shapiro CL, Lin HJ, Cheng AS, Jin VX, Huang TH',
'description' => 'Trimethylation of histone 3 lysine 27 (H3K27me3) is a critical epigenetic mark for the maintenance of gene silencing. Additional accumulation of DNA methylation in target loci is thought to cooperatively support this epigenetic silencing during tumorigenesis. However, molecular mechanisms underlying the complex interplay between the two marks remain to be explored. Here we show that activation of PI3K/AKT signaling can be a trigger of this epigenetic processing at many downstream target genes. We also find that DNA methylation can be acquired at the same loci in cancer cells, thereby reinforcing permanent repression in those losing the H3K27me3 mark. Because of a link between PI3K/AKT signaling and epigenetic alterations, we conducted epigenetic therapies in conjunction with the signaling-targeted treatment. These combined treatments synergistically relieve gene silencing and suppress cancer cell growth in vitro and in xenografts. The new finding has important implications for improving targeted cancer therapies in the future.',
'date' => '2011-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/21216892',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 105 => array(
'id' => '412',
'name' => 'Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping.',
'authors' => 'Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, Weng YI, Deatherage DE, Chen Z, Pereira JS, Lopez R, Russo J, Wang Q, Lamartiniere CA, Nephew KP, Huang TH',
'description' => 'The current concept of epigenetic repression is based on one repressor unit corresponding to one silent gene. This notion, however, cannot adequately explain concurrent silencing of multiple loci observed in large chromosome regions. The long-range epigenetic silencing (LRES) can be a frequent occurrence throughout the human genome. To comprehensively characterize the influence of estrogen signaling on LRES, we analyzed transcriptome, methylome, and estrogen receptor alpha (ESR1)-binding datasets from normal breast epithelia and breast cancer cells. This "omics" approach uncovered 11 large repressive zones (range, 0.35 approximately 5.98 megabases), including a 14-gene cluster located on 16p11.2. In normal cells, estrogen signaling induced transient formation of multiple DNA loops in the 16p11.2 region by bringing 14 distant loci to focal ESR1-docking sites for coordinate repression. However, the plasticity of this free DNA movement was reduced in breast cancer cells. Together with the acquisition of DNA methylation and repressive chromatin modifications at the 16p11.2 loci, an inflexible DNA scaffold may be a novel determinant used by breast cancer cells to reinforce estrogen-mediated repression.',
'date' => '2010-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/20442245',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 106 => array(
'id' => '976',
'name' => 'Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells.',
'authors' => 'Borgel J, Guibert S, Weber M.',
'description' => 'Methylated DNA immunoprecipitation (MeDIP) is an immunocapturing approach for unbiased enrichment of DNA that is methylated on cytosines. The principle is that genomic DNA is randomly sheared by sonication and immunoprecipitated with an antibody that specifically recognizes 5-methylcytidine (5mC), which can be combined with PCR or high-throughput analysis (microarrays, deep sequencing). The MeDIP technique has been originally used to generate DNA methylation profiles on a genome scale in mammals and plants. Here we provide an optimized version of the MeDIP protocol suitable for low amounts of DNA, which can be used to study DNA methylation in cellular populations available in small quantities.',
'date' => '0000-00-00',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22907495',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '120',
'name' => '5-mC Antibody - clone 33D3 SDS US en',
'language' => 'en',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-US-en-GHS_2_0.pdf',
'countries' => 'US',
'modified' => '2020-03-13 15:47:06',
'created' => '2020-03-13 15:47:06',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '118',
'name' => '5-mC Antibody - clone 33D3 SDS GB en',
'language' => 'en',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-GB-en-GHS_2_0.pdf',
'countries' => 'GB',
'modified' => '2020-03-13 15:44:22',
'created' => '2020-03-13 15:44:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '116',
'name' => '5-mC Antibody - clone 33D3 SDS ES es',
'language' => 'es',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-ES-es-GHS_2_0.pdf',
'countries' => 'ES',
'modified' => '2020-03-13 15:43:01',
'created' => '2020-03-13 15:43:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '115',
'name' => '5-mC Antibody - clone 33D3 SDS DE de',
'language' => 'de',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-DE-de-GHS_2_0.pdf',
'countries' => 'DE',
'modified' => '2020-03-13 15:42:22',
'created' => '2020-03-13 15:42:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '119',
'name' => '5-mC Antibody - clone 33D3 SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-JP-ja-GHS_4_0.pdf',
'countries' => 'JP',
'modified' => '2020-03-13 15:44:56',
'created' => '2020-03-13 15:44:56',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '114',
'name' => '5-mC Antibody - clone 33D3 SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-03-13 15:41:00',
'created' => '2020-03-13 15:41:00',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '113',
'name' => '5-mC Antibody - clone 33D3 SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-BE-fr-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-03-13 15:40:12',
'created' => '2020-03-13 15:40:12',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '117',
'name' => '5-mC Antibody - clone 33D3 SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-03-13 15:43:48',
'created' => '2020-03-13 15:43:48',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = true
$other_formats = array(
(int) 0 => array(
'id' => '1980',
'antibody_id' => '630',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>Monoclonal antibody raised in mouse against </span><b>5-mC</b><span><span> </span>(</span><b>5-methylcytosine</b><span>) conjugated to ovalbumine (</span><b>33D3 clone</b><span>).</span></p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15200081-100',
'old_catalog_number' => 'MAb-081-100',
'sf_code' => 'C15200081-D001-000526',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '505',
'price_USD' => '575',
'price_GBP' => '450',
'price_JPY' => '79110',
'price_CNY' => '0',
'price_AUD' => '1438',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'October 27, 2020',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081) | Diagenode',
'meta_keywords' => '5-methylcytosine (5-mC),monoclonal antibody,Methylated DNA Immunoprecipitation',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2023-05-17 10:08:33',
'created' => '2015-06-29 14:08:20'
),
(int) 1 => array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
)
$pro = array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010034</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1885" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1885" id="CartAdd/1885Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1885" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto hMeDIP kit x16 (monoclonal mouse antibody)</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns" data-reveal-id="cartModal-1885" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto hMeDIP kit x16 (monoclonal mouse antibody)</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/magmedip-kit-x48-48-rxns"><img src="/img/product/kits/C02010021-magmedip-qpcr.jpg" alt="MagMeDIP qPCR Kit box" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010021</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1880" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1880" id="CartAdd/1880Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1880" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MagMeDIP Kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MagMeDIP Kit',
'C02010021',
'750',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MagMeDIP Kit',
'C02010021',
'750',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="magmedip-kit-x48-48-rxns" data-reveal-id="cartModal-1880" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MagMeDIP Kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/auto-methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020011</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1888" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1888" id="CartAdd/1888Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1888" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto MethylCap kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1888" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto MethylCap kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1887" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1887" id="CartAdd/1887Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1887" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MethylCap kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MethylCap kit',
'C02020010',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MethylCap kit',
'C02020010',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1887" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MethylCap kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/premium-bisulfite-kit-50-rxns"><img src="/img/grey-logo.jpg" alt="default alt" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02030030</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1892" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1892" id="CartAdd/1892Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1892" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Premium Bisulfite kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Premium Bisulfite kit',
'C02030030',
'240',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Premium Bisulfite kit',
'C02030030',
'240',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="premium-bisulfite-kit-50-rxns" data-reveal-id="cartModal-1892" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Premium Bisulfite kit</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1892',
'antibody_id' => null,
'name' => 'Premium Bisulfite kit',
'description' => '<p style="text-align: center;"><a href="https://www.diagenode.com/files/products/kits/Premium_Bisulfite_kit_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p style="text-align: center;"><strong>Make your Bisulfite conversion now in only 60 minutes !</strong></p>
<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C02030030',
'old_catalog_number' => '',
'sf_code' => 'C02030030-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'premium-bisulfite-kit-50-rxns',
'meta_title' => 'Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Premium Bisulfite kit',
'modified' => '2023-04-20 16:13:50',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '271',
'product_id' => '1979',
'related_id' => '1892'
),
'Image' => array()
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ' <span style="color:#CCC">(MAb-081-500)</span>'
$country_code = 'US'
$other_format = array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
'id' => '1914',
'product_id' => '1979',
'application_id' => '29'
)
)
$slugs = array(
(int) 0 => 'immunofluorescence'
)
$applications = array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'locale' => 'eng'
)
$description = '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>'
$name = 'IF'
$document = array(
'id' => '1118',
'name' => 'Datasheet 5-mC33D3 C15200081-10',
'description' => '<p><span>The 5-methylcytosine antibody (clone 33D3) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span></p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_5-mC33D3_C15200081-10.pdf',
'slug' => 'datasheet-5-mc33d3-c15200081-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-12-17 10:55:09',
'created' => '2020-12-17 10:55:09',
'ProductsDocument' => array(
'id' => '3063',
'product_id' => '1979',
'document_id' => '1118'
)
)
$sds = array(
'id' => '117',
'name' => '5-mC Antibody - clone 33D3 SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-03-13 15:43:48',
'created' => '2020-03-13 15:43:48',
'ProductsSafetySheet' => array(
'id' => '231',
'product_id' => '1979',
'safety_sheet_id' => '117'
)
)
$publication = array(
'id' => '976',
'name' => 'Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells.',
'authors' => 'Borgel J, Guibert S, Weber M.',
'description' => 'Methylated DNA immunoprecipitation (MeDIP) is an immunocapturing approach for unbiased enrichment of DNA that is methylated on cytosines. The principle is that genomic DNA is randomly sheared by sonication and immunoprecipitated with an antibody that specifically recognizes 5-methylcytidine (5mC), which can be combined with PCR or high-throughput analysis (microarrays, deep sequencing). The MeDIP technique has been originally used to generate DNA methylation profiles on a genome scale in mammals and plants. Here we provide an optimized version of the MeDIP protocol suitable for low amounts of DNA, which can be used to study DNA methylation in cellular populations available in small quantities.',
'date' => '0000-00-00',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22907495',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
'id' => '711',
'product_id' => '1979',
'publication_id' => '976'
)
)
$externalLink = ' <a href="http://www.ncbi.nlm.nih.gov/pubmed/22907495" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: message [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'product' => array(
'Product' => array(
'id' => '1979',
'antibody_id' => '631',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (sample size)',
'description' => '<p>Monoclonal antibody raised in mouse against 5-mC (<strong>5-methylcytosine</strong>) conjugated to ovalbumine (<strong>33D3 clone</strong>).</p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15200081-10',
'old_catalog_number' => 'MAb-081-010',
'sf_code' => 'C15200081-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10-ug-5-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:58:36',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '631',
'name' => '5-mC monoclonal antibody 33D3 (sample size)',
'description' => '',
'clonality' => '',
'isotype' => 'IgG1',
'lot' => 'RD-006',
'concentration' => '1.1 µg/µl',
'reactivity' => 'Human, mouse, rat, cow, other (wide range): positive',
'type' => 'Monoclonal. <br /><strong>MeDIP grade.</strong><br /><strong>MeDIP-seq grade</strong>.',
'purity' => 'Protein A purified monoclonal antibody.',
'classification' => '',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>MeDIP/MeDIP-seq <sup>*</sup></td>
<td>0.1 - 2 µg/IP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting **</td>
<td>1:300</td>
<td>Fig 3</td>
</tr>
<tr>
<td>IF</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
</tbody>
</table>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.1-5 µg per IP.</small></p>
<p><small><sup>**</sup> Dot blot was only performed to demonstrate the specificity. This antibody is not recommended for dot blot on biological samples.</small></p>
<p></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2023-05-11 15:21:00',
'created' => '2020-12-17 10:31:21',
'select_label' => '631 - 5-mC monoclonal antibody 33D3 (sample size) (RD-006 - 1.1 µg/µl - Human, mouse, rat, cow, other (wide range): positive - Protein A purified monoclonal antibody. - Mouse)'
),
'Slave' => array(),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
),
(int) 25 => array(
[maximum depth reached]
),
(int) 26 => array(
[maximum depth reached]
),
(int) 27 => array(
[maximum depth reached]
),
(int) 28 => array(
[maximum depth reached]
),
(int) 29 => array(
[maximum depth reached]
),
(int) 30 => array(
[maximum depth reached]
),
(int) 31 => array(
[maximum depth reached]
),
(int) 32 => array(
[maximum depth reached]
),
(int) 33 => array(
[maximum depth reached]
),
(int) 34 => array(
[maximum depth reached]
),
(int) 35 => array(
[maximum depth reached]
),
(int) 36 => array(
[maximum depth reached]
),
(int) 37 => array(
[maximum depth reached]
),
(int) 38 => array(
[maximum depth reached]
),
(int) 39 => array(
[maximum depth reached]
),
(int) 40 => array(
[maximum depth reached]
),
(int) 41 => array(
[maximum depth reached]
),
(int) 42 => array(
[maximum depth reached]
),
(int) 43 => array(
[maximum depth reached]
),
(int) 44 => array(
[maximum depth reached]
),
(int) 45 => array(
[maximum depth reached]
),
(int) 46 => array(
[maximum depth reached]
),
(int) 47 => array(
[maximum depth reached]
),
(int) 48 => array(
[maximum depth reached]
),
(int) 49 => array(
[maximum depth reached]
),
(int) 50 => array(
[maximum depth reached]
),
(int) 51 => array(
[maximum depth reached]
),
(int) 52 => array(
[maximum depth reached]
),
(int) 53 => array(
[maximum depth reached]
),
(int) 54 => array(
[maximum depth reached]
),
(int) 55 => array(
[maximum depth reached]
),
(int) 56 => array(
[maximum depth reached]
),
(int) 57 => array(
[maximum depth reached]
),
(int) 58 => array(
[maximum depth reached]
),
(int) 59 => array(
[maximum depth reached]
),
(int) 60 => array(
[maximum depth reached]
),
(int) 61 => array(
[maximum depth reached]
),
(int) 62 => array(
[maximum depth reached]
),
(int) 63 => array(
[maximum depth reached]
),
(int) 64 => array(
[maximum depth reached]
),
(int) 65 => array(
[maximum depth reached]
),
(int) 66 => array(
[maximum depth reached]
),
(int) 67 => array(
[maximum depth reached]
),
(int) 68 => array(
[maximum depth reached]
),
(int) 69 => array(
[maximum depth reached]
),
(int) 70 => array(
[maximum depth reached]
),
(int) 71 => array(
[maximum depth reached]
),
(int) 72 => array(
[maximum depth reached]
),
(int) 73 => array(
[maximum depth reached]
),
(int) 74 => array(
[maximum depth reached]
),
(int) 75 => array(
[maximum depth reached]
),
(int) 76 => array(
[maximum depth reached]
),
(int) 77 => array(
[maximum depth reached]
),
(int) 78 => array(
[maximum depth reached]
),
(int) 79 => array(
[maximum depth reached]
),
(int) 80 => array(
[maximum depth reached]
),
(int) 81 => array(
[maximum depth reached]
),
(int) 82 => array(
[maximum depth reached]
),
(int) 83 => array(
[maximum depth reached]
),
(int) 84 => array(
[maximum depth reached]
),
(int) 85 => array(
[maximum depth reached]
),
(int) 86 => array(
[maximum depth reached]
),
(int) 87 => array(
[maximum depth reached]
),
(int) 88 => array(
[maximum depth reached]
),
(int) 89 => array(
[maximum depth reached]
),
(int) 90 => array(
[maximum depth reached]
),
(int) 91 => array(
[maximum depth reached]
),
(int) 92 => array(
[maximum depth reached]
),
(int) 93 => array(
[maximum depth reached]
),
(int) 94 => array(
[maximum depth reached]
),
(int) 95 => array(
[maximum depth reached]
),
(int) 96 => array(
[maximum depth reached]
),
(int) 97 => array(
[maximum depth reached]
),
(int) 98 => array(
[maximum depth reached]
),
(int) 99 => array(
[maximum depth reached]
),
(int) 100 => array(
[maximum depth reached]
),
(int) 101 => array(
[maximum depth reached]
),
(int) 102 => array(
[maximum depth reached]
),
(int) 103 => array(
[maximum depth reached]
),
(int) 104 => array(
[maximum depth reached]
),
(int) 105 => array(
[maximum depth reached]
),
(int) 106 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul'
)
$language = 'en'
$meta_keywords = ''
$meta_description = '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.'
$meta_title = '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode'
$product = array(
'Product' => array(
'id' => '1979',
'antibody_id' => '631',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (sample size)',
'description' => '<p>Monoclonal antibody raised in mouse against 5-mC (<strong>5-methylcytosine</strong>) conjugated to ovalbumine (<strong>33D3 clone</strong>).</p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15200081-10',
'old_catalog_number' => 'MAb-081-010',
'sf_code' => 'C15200081-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10-ug-5-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:58:36',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '631',
'name' => '5-mC monoclonal antibody 33D3 (sample size)',
'description' => '',
'clonality' => '',
'isotype' => 'IgG1',
'lot' => 'RD-006',
'concentration' => '1.1 µg/µl',
'reactivity' => 'Human, mouse, rat, cow, other (wide range): positive',
'type' => 'Monoclonal. <br /><strong>MeDIP grade.</strong><br /><strong>MeDIP-seq grade</strong>.',
'purity' => 'Protein A purified monoclonal antibody.',
'classification' => '',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>MeDIP/MeDIP-seq <sup>*</sup></td>
<td>0.1 - 2 µg/IP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting **</td>
<td>1:300</td>
<td>Fig 3</td>
</tr>
<tr>
<td>IF</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
</tbody>
</table>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.1-5 µg per IP.</small></p>
<p><small><sup>**</sup> Dot blot was only performed to demonstrate the specificity. This antibody is not recommended for dot blot on biological samples.</small></p>
<p></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2023-05-11 15:21:00',
'created' => '2020-12-17 10:31:21',
'select_label' => '631 - 5-mC monoclonal antibody 33D3 (sample size) (RD-006 - 1.1 µg/µl - Human, mouse, rat, cow, other (wide range): positive - Protein A purified monoclonal antibody. - Mouse)'
),
'Slave' => array(),
'Group' => array(
'Group' => array(
'id' => '4',
'name' => 'C15200081',
'product_id' => '1980',
'modified' => '2016-02-17 17:32:42',
'created' => '2016-02-17 17:32:42'
),
'Master' => array(
'id' => '1980',
'antibody_id' => '630',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>Monoclonal antibody raised in mouse against </span><b>5-mC</b><span><span> </span>(</span><b>5-methylcytosine</b><span>) conjugated to ovalbumine (</span><b>33D3 clone</b><span>).</span></p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15200081-100',
'old_catalog_number' => 'MAb-081-100',
'sf_code' => 'C15200081-D001-000526',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '505',
'price_USD' => '575',
'price_GBP' => '450',
'price_JPY' => '79110',
'price_CNY' => '0',
'price_AUD' => '1438',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'October 27, 2020',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081) | Diagenode',
'meta_keywords' => '5-methylcytosine (5-mC),monoclonal antibody,Methylated DNA Immunoprecipitation',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2023-05-17 10:08:33',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1885',
'antibody_id' => null,
'name' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'description' => '<p><span>The Auro hMeDIP kit is designed for enrichment of hydroxymethylated DNA from fragmented genomic DNA samples for use in genome-wide methylation analysis. It features</span><span> a highly specific monoclonal antibody against </span><span>5-hydroxymethylcytosine (5-hmC) for the immunoprecipitation of hydroxymethylated DNA</span><span>. It includes control DNA and primers to assess the effiency of the assay. </span><span>Performing hydroxymethylation profiling with the hMeDIP kit is fast, reliable and highly specific.</span></p>',
'label1' => ' Characteristics',
'info1' => '<ul style="list-style-type: disc;">
<li><span>Robust enrichment & immunoprecipitation of hydroxymethylated DNA</span></li>
<li>Highly specific monoclonal antibody against 5-hmC<span> for reliable, reproducible results</span></li>
<li>Including control DNA and primers to <span>monitor the efficiency of the assay</span>
<ul style="list-style-type: circle;">
<li>5-hmC, 5-mC and unmethylated DNA sequences and primer pairs</li>
<li>Mouse primer pairs against Sfi1 targeting hydroxymethylated gene in mouse</li>
</ul>
</li>
</ul>
<ul style="list-style-type: disc;">
<li>Improved single-tube, magnetic bead-based protocol</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C02010034',
'old_catalog_number' => '',
'sf_code' => 'C02010034-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '630',
'price_USD' => '690',
'price_GBP' => '580',
'price_JPY' => '98690',
'price_CNY' => '',
'price_AUD' => '1725',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns',
'meta_title' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'meta_keywords' => '',
'meta_description' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'modified' => '2021-01-18 10:37:19',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1880',
'antibody_id' => null,
'name' => 'MagMeDIP Kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/magmedip-kit-manual-C02010020-21.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p> </p>
<div class="small-12 medium-4 large-4 columns"><center></center><center></center><center></center><center><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank"><img src="https://www.diagenode.com/img/banners/banner-nature-publication-580.png" alt="Click here to read more about MeDIP " caption="false" width="80%" /></a></center></div>
<div class="small-12 medium-8 large-8 columns">
<h3 style="text-align: justify;">Sensitive tumour detection and classification using plasma cell-free DNA methylomes<br /><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank">Read the publication</a></h3>
<h3 class="c-article-title u-h1" data-test="article-title" itemprop="name headline" style="text-align: justify;">Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA<br /><a href="https://www.nature.com/articles/s41596-019-0202-2" target="_blank" title="cfMeDIP-seq Nature Method">Read the method</a></h3>
</div>
<p></p>
<p></p>
<p></p>
<div class="row">
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
<div class="small-12 medium-8 large-8 columns"><br />
<p>Perform <strong>MeDIP</strong> (<strong>Me</strong>thylated <strong>D</strong>NA <strong>I</strong>mmuno<strong>p</strong>recipitation) followed by qPCR or NGS to estimate DNA methylation status of your sample using a highly sensitive 5-methylcytosine antibody. Our MagMeDIP kit contains high quality reagents to get the highest enrichment of methylated DNA with an optimized user-friendly protocol.</p>
</div>
</div>
<h3><span>Features</span></h3>
<ul>
<li>Starting DNA amount: <strong>10 ng – 1 µg</strong></li>
<li>Content: <strong>all reagents included</strong> for DNA extraction, immunoprecipitation (including the 5-mC antibody, spike-in controls and their corresponding qPCR primer pairs) as well as DNA isolation after IP.</li>
<li>Application: <strong>qPCR</strong> and <strong>NGS</strong></li>
<li>Robust method, <strong>superior enrichment</strong>, and easy-to-use protocol</li>
<li><strong>High reproducibility</strong> between replicates and repetitive experiments</li>
<li>Compatible with <strong>all species </strong></li>
</ul>',
'label1' => 'MagMeDIP workflow',
'info1' => '<p>DNA methylation occurs primarily as 5-methylcytosine (5-mC), and the Diagenode MagMeDIP Kit takes advantage of a specific antibody targeting this 5-mC to immunoprecipitate methylated DNA, which can be thereafter directly analyzed by qPCR or Next-Generation Sequencing (NGS).</p>
<h3><span>How it works</span></h3>
<p>In brief, after the cell collection and lysis, the genomic DNA is extracted, sheared, and then denatured. In the next step the antibody directed against 5 methylcytosine and antibody binding beads are used for immunoselection and immunoprecipitation of methylated DNA fragments. Then, the IP’d methylated DNA is isolated and can be used for any subsequent analysis as qPCR, amplification, hybridization on microarrays or next generation sequencing.</p>
<center><img src="https://www.diagenode.com/img/product/kits/MagMeDIP-workflow.png" width="70%" alt="5-methylcytosine" caption="false" /></center>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>',
'label2' => 'MeDIP-qPCR',
'info2' => '<p>The kit MagMeDIP contains all reagents necessary for a complete MeDIP-qPCR workflow. Two MagMeDIP protocols have been validated: for manual processing as well as for automated processing, using the Diagenode’s IP-Star Compact Automated System (please refer to the kit manual).</p>
<ul>
<li><strong>Complete kit</strong> including DNA extraction module, IP antibody and reagents, DNA isolation buffer</li>
<li><strong>Quality control of the IP:</strong> due to methylated and unmethylated DNA spike-in controls and their associated qPCR primers</li>
<li><strong>Easy to use</strong> with user-friendly magnetic beads and rack</li>
<li><strong>Highly validated protocol</strong></li>
<li>Automated protocol supplied</li>
</ul>
<center><img src="https://www.diagenode.com/img/product/kits/fig1-magmedipkit.png" width="85%" alt="Methylated DNA Immunoprecipitation" caption="false" /></center>
<p style="font-size: 0.9em;"><em><strong>Figure 1.</strong> Immunoprecipitation results obtained with Diagenode MagMeDIP Kit</em></p>
<p style="font-size: 0.9em;">MeDIP assays were performed manually using 1 µg or 50 ng gDNA from blood cells with the MagMeDIP kit (Diagenode). The IP was performed with the Methylated and Unmethylated spike-in controls included in the kit, together with the human DNA samples. The DNA was isolated/purified using DIB. Afterwards, qPCR was performed using the primer pairs included in this kit.</p>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>',
'label3' => 'MeDIP-seq',
'info3' => '<p>For DNA methylation analysis on the whole genome, MagMeDIP kit can be coupled with Next-Generation Sequencing. To perform MeDIP-sequencing we recommend the following strategy:</p>
<ul style="list-style-type: circle;">
<li>Choose a library preparation solution which is compatible with the starting amount of DNA you are planning to use (from 10 ng to 1 μg). It can be a home-made solution or a commercial one.</li>
<li>Choose the indexing system that fits your needs considering the following features:</li>
<ul>
<ul>
<ul>
<li>Single-indexing, combinatorial dual-indexing or unique dual-indexing</li>
<li>Number of barcodes</li>
<li>Full-length adaptors containing the barcodes or barcoding at the final amplification step</li>
<li>Presence / absence of Unique Molecular Identifiers (for PCR duplicates removal)</li>
</ul>
</ul>
</ul>
<li>Standard library preparation protocols are compatible with double-stranded DNA only, therefore the first steps of the library preparation (end repair, A-tailing, adaptor ligation and clean-up) will have to be performed on sheared DNA, before the IP.</li>
</ul>
<p style="padding-left: 30px;"><strong>CAUTION:</strong> As the immunoprecipitation step occurs at the middle of the library preparation workflow, single-tube solutions for library preparation are usually not compatible with MeDIP-sequencing.</p>
<ul style="list-style-type: circle;">
<li>For DNA isolation after the IP, we recommend using the <a href="https://www.diagenode.com/en/p/ipure-kit-v2-x24" title="IPure kit v2">IPure kit v2</a> (available separately, Cat. No. C03010014) instead of DNA isolation Buffer.</li>
</ul>
<ul style="list-style-type: circle;">
<li>Perform library amplification after the DNA isolation following the standard protocol of the chosen library preparation solution.</li>
</ul>
<h3><span>MeDIP-seq workflow</span></h3>
<center><img src="https://www.diagenode.com/img/product/kits/MeDIP-seq-workflow.png" width="110%" alt="MagMeDIP qPCR Kit x10 workflow" caption="false" /></center>
<h3><span>Example of results</span></h3>
<center><img src="https://www.diagenode.com/img/product/kits/medip-specificity.png" alt="MagMeDIP qPCR Kit Result" caption="false" width="951" height="488" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 1. qPCR analysis of external spike-in DNA controls (methylated and unmethylated) after IP.</strong> Samples were prepared using 1μg – 100ng -10ng sheared human gDNA with the MagMeDIP kit (Diagenode) and a commercially available library prep kit. DNA isolation after IP has been performed with IPure kit V2 (Diagenode).</p>
<p></p>
<p></p>
<center><img src="https://www.diagenode.com/img/product/kits/medip-saturation-analysis.png" alt=" MagMeDIP kit " caption="false" width="951" height="461" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 2. Saturation analysis.</strong> Clean reads were aligned to the human genome (hg19) using Burrows-Wheeler aligner (BWA) algorithm after which duplicated and unmapped reads were removed resulting in a mapping efficiency >98% for all samples. Quality and validity check of the mapped MeDIP-seq data was performed using MEDIPS R package. Saturation plots show that all sets of reads have sufficient complexity and depth to saturate the coverage profile of the reference genome and that this is reproducible between replicates and repetitive experiments (data shown for 50 ng gDNA input: left panel = replicate a, right panel = replicate b).</p>
<p></p>
<p></p>
<center><img src="https://www.diagenode.com/img/product/kits/medip-libraries-prep.png" alt="MagMeDIP x10 " caption="false" width="951" height="708" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 3. Sequencing profiles of MeDIP-seq libraries prepared from different starting amounts of sheared gDNA on the positive and negative methylated control regions.</strong> MeDIP-seq libraries were prepared from decreasing starting amounts of gDNA (1 μg (green), 50 ng (red), and 10ng (blue)) originating from human blood with the MagMeDIP kit (Diagenode) and a commercially available library prep kit. DNA isolation after IP has been performed with IPure kit V2 (Diagenode). IP and corresponding INPUT samples were sequenced on Illumina NovaSeq SP with 2x50 PE reads. The reads were mapped to the human genome (hg19) with bwa and the alignments were loaded into IGV (the tracks use an identical scale). The top IGV figure shows the TSH2B (also known as H2BC1) gene (marked by blue boxes in the bottom track) and its surroundings. The TSH2B gene is coding for a histone variant that does not occur in blood cells, and it is known to be silenced by methylation. Accordingly, we see a high coverage in the vicinity of this gene. The bottom IGV figure shows the GADPH locus (marked by blue boxes in the bottom track) and its surroundings. The GADPH gene is a highly active transcription region and should not be methylated, resulting in no reads accumulation following MeDIP-seq experiment.</p>
<p></p>
<ul>
<ul>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
</ul>
</ul>',
'format' => '48 rxns (IP)',
'catalog_number' => 'C02010021',
'old_catalog_number' => 'mc-magme-048',
'sf_code' => 'C02010021-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '745',
'price_USD' => '750',
'price_GBP' => '680',
'price_JPY' => '116705',
'price_CNY' => '',
'price_AUD' => '1875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'magmedip-kit-x48-48-rxns',
'meta_title' => 'MagMeDIP Kit for efficient immunoprecipitation of methylated DNA | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Perform Methylated DNA Immunoprecipitation (MeDIP) to estimate DNA methylation status of your sample using highly specific 5-mC antibody. This kit allows the preparation of cfMeDIP-seq libraries.',
'modified' => '2023-07-06 14:15:10',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1887',
'antibody_id' => null,
'name' => 'MethylCap kit',
'description' => '<p>The MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. The procedure has been adapted to both manual process or <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a>. Libraries of captured methylated DNA can be prepared for next-generation sequencing (NGS) by combining MBD technology with the <a href="https://www.diagenode.com/en/p/microplex-lib-prep-kit-v3-48-rxns">MicroPlex Library Preparation Kit v3</a>.</p>',
'label1' => 'Characteristics',
'info1' => '<ul style="list-style-type: circle;">
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>On-day protocol</strong></li>
<li><strong>NGS compatibility</strong></li>
</ul>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong></strong></p>
<p><strong></strong><strong>F</strong><strong>igure 1.</strong> Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).</p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020010',
'old_catalog_number' => 'AF-100-0048',
'sf_code' => 'C02020010-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'methylcap-kit-x48-48-rxns',
'meta_title' => 'MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'MethylCap kit x48',
'modified' => '2021-02-19 17:55:38',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '1892',
'antibody_id' => null,
'name' => 'Premium Bisulfite kit',
'description' => '<p style="text-align: center;"><a href="https://www.diagenode.com/files/products/kits/Premium_Bisulfite_kit_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p style="text-align: center;"><strong>Make your Bisulfite conversion now in only 60 minutes !</strong></p>
<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C02030030',
'old_catalog_number' => '',
'sf_code' => 'C02030030-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'premium-bisulfite-kit-50-rxns',
'meta_title' => 'Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Premium Bisulfite kit',
'modified' => '2023-04-20 16:13:50',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array([maximum depth reached])
)
),
'Application' => array(
(int) 0 => array(
'id' => '28',
'position' => '10',
'parent_id' => '40',
'name' => 'DB',
'description' => '<p>Dot blotting</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'dot-blotting',
'meta_keywords' => 'Dot blotting,Monoclonal & Polyclonal antibody,',
'meta_description' => 'Diagenode offers Monoclonal & Polyclonal antibodies for Dot blotting applications',
'meta_title' => 'Dot blotting Antibodies - Monoclonal & Polyclonal antibody | Diagenode',
'modified' => '2016-01-13 14:40:49',
'created' => '2015-07-08 13:45:05',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '23',
'position' => '40',
'parent_id' => '4',
'name' => 'DNA modifications',
'description' => '<p><span style="font-weight: 400;">T</span><span style="font-weight: 400;">he pattern of <strong>DNA modifications</strong> is critical for genome stability and the control of gene expression in the cell. Methylation of 5-cytosine (5-mC), one of the best-studied epigenetic marks, is carried out by the <strong>DNA methyltransferases</strong> DNMT3A and B and DNMT1. DNMT3A and DNMT3B are responsible for </span><i><span style="font-weight: 400;">de novo</span></i><span style="font-weight: 400;"> DNA methylation, whereas DNMT1 maintains existing methylation. 5-mC undergoes active demethylation which is performed by the <strong>Ten-Eleven Translocation</strong> (TET) familly of DNA hydroxylases. The latter consists of 3 members TET1, 2 and 3. All 3 members catalyze the conversion of <strong>5-methylcytosine</strong> (5-mC) into <strong>5-hydroxymethylcytosine</strong> (5-hmC), and further into <strong>5-formylcytosine</strong> (5-fC) and <strong>5-carboxycytosine</strong> (5-caC). 5-fC and 5-caC can be converted to unmodified cytosine by <strong>Thymine DNA Glycosylase</strong> (TDG). It is not yet clear if 5-hmC, 5-fC and 5-caC have specific functions or are simply intermediates in the demethylation of 5-mC.</span></p>
<p><span style="font-weight: 400;">DNA methylation is generally considered as a repressive mark and is usually associated with gene silencing. It is essential that the balance between DNA methylation and demethylation is precisely maintained. Dysregulation of DNA methylation may lead to many different human diseases and is often observed in cancer cells.</span></p>
<p><span style="font-weight: 400;">Diagenode offers highly validated antibodies against different proteins involved in DNA modifications as well as against the modified bases allowing the study of all steps and intermediates in the DNA methylation/demethylation pathway:</span></p>
<p><img src="https://www.diagenode.com/img/categories/antibodies/dna-methylation.jpg" height="599" width="816" /></p>
<p><strong>Diagenode exclusively sources the original 5-methylcytosine monoclonal antibody (clone 33D3).</strong></p>
<p>Check out the list below to see all proposed antibodies for DNA modifications.</p>
<p>Diagenode’s highly validated antibodies:</p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'dna-methylation-antibody',
'cookies_tag_id' => null,
'meta_keywords' => 'Monoclonal Antibodies,Polyclonal antibodies,DNA methylation,Diagenode',
'meta_description' => 'Diagenode offers Monoclonal and Polyclonal antibodies for DNA Methylation. The pattern of DNA modifications is critical for genome stability and the control of gene expression in the cell. ',
'meta_title' => 'DNA modifications - Monoclonal and Polyclonal Antibodies for DNA Methylation | Diagenode',
'modified' => '2020-08-03 15:25:37',
'created' => '2014-07-31 03:51:53',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '102',
'position' => '1',
'parent_id' => '4',
'name' => 'Sample size antibodies',
'description' => '<h1><strong>Validated epigenetics antibodies</strong> – care for a sample?<br /> </h1>
<p>Diagenode has partnered with leading epigenetics experts and numerous epigenetics consortiums to bring to you a validated and comprehensive collection of epigenetic antibodies. As an expert in epigenetics, we are committed to offering highly-specific antibodies validated for ChIP/ChIP-seq and many other applications. All batch-specific validation data is available on our website.<br /><a href="../categories/antibodies">Read about our expertise in antibody production</a>.</p>
<ul>
<li><strong>Focused</strong> - Diagenode's selection of antibodies is exclusively dedicated for epigenetic research. <a title="See the full collection." href="../categories/all-antibodies">See the full collection.</a></li>
<li><strong>Strict quality standards</strong> with rigorous QC and validation</li>
<li><strong>Classified</strong> based on level of validation for flexibility of application</li>
</ul>
<p>Existing sample sizes are listed below. We will soon expand our collection. Are you looking for a sample size of another antibody? Just <a href="mailto:agnieszka.zelisko@diagenode.com?Subject=Sample%20Size%20Request" target="_top">Contact us</a>.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => true,
'is_antibody' => true,
'slug' => 'sample-size-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => '5-hmC monoclonal antibody,CRISPR/Cas9 polyclonal antibody ,H3K36me3 polyclonal antibody,diagenode',
'meta_description' => 'Diagenode offers sample volume on selected antibodies for researchers to test, validate and provide confidence and flexibility in choosing from our wide range of antibodies ',
'meta_title' => 'Sample-size Antibodies | Diagenode',
'modified' => '2019-07-03 10:57:05',
'created' => '2015-10-27 12:13:34',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '15',
'name' => 'Monoclonal Antibody hMeDIP Kit for DNA Hydroxymethylation Studies',
'description' => '<p><span>There is substantial interest and speculation in the role of the “sixth DNA base,” 5-hydroxymethylcytosine (5-hmC), although its precise function has not yet been elucidated. Since its discovery in neuronal Purkinje, granule and ES cells, studies of this new modified DNA base have been limited by the lack of high-quality, validated tools and technologies that discriminate hydroxy-methylation from methylation in regulating genome expression. Obtaining a specific assay for 5-hmC is particularly important since standard bisulfite sequencing cannot distinguish between these two types of methylation. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Monoclonal_Antibody_hMeDIP_Kit_for_DNA_Hydroxymethylation_Studies_Poster.pdf',
'slug' => 'monoclonal-antibody-hmedip-kit-for-dna-hydroxymethylation-studies-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-09-29 13:42:52',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1118',
'name' => 'Datasheet 5-mC33D3 C15200081-10',
'description' => '<p><span>The 5-methylcytosine antibody (clone 33D3) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span></p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_5-mC33D3_C15200081-10.pdf',
'slug' => 'datasheet-5-mc33d3-c15200081-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-12-17 10:55:09',
'created' => '2020-12-17 10:55:09',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '250',
'name' => 'product/antibodies/antibody.png',
'alt' => 'Mouse IgG',
'modified' => '2020-11-27 07:00:09',
'created' => '2015-07-17 10:12:18',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4843',
'name' => 'Differentiation block in acute myeloid leukemia regulated by intronicsequences of FTO',
'authors' => 'Camera F. et al.',
'description' => '<p>Iroquois transcription factor gene IRX3 is highly expressed in 20–30\% of acute myeloid leukemia (AML) and contributes to the pathognomonic differentiation block. Intron 8 FTO sequences ∼220kB downstream of IRX3 exhibit histone acetylation, DNA methylation, and contacts with the IRX3 promoter, which correlate with IRX3 expression. Deletion of these intronic elements confirms a role in positively regulating IRX3. RNAseq revealed long non-coding (lnc) transcripts arising from this locus. FTO-lncAML knockdown (KD) induced differentiation of AML cells, loss of clonogenic activity, and reduced FTO intron 8:IRX3 promoter contacts. While both FTO-lncAML KD and IRX3 KD induced differentiation, FTO-lncAML but not IRX3 KD led to HOXA downregulation suggesting transcript activity in trans. FTO-lncAMLhigh AML samples expressed higher levels of HOXA and lower levels of differentiation genes. Thus, a regulatory module in FTO intron 8 consisting of clustered enhancer elements and a long non-coding RNA is active in human AML, impeding myeloid differentiation.</p>',
'date' => '2023-08-01',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2589004223013962',
'doi' => '10.1016/j.isci.2023.107319',
'modified' => '2023-08-01 14:14:01',
'created' => '2023-08-01 15:59:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4796',
'name' => 'Nicotinamide N-methyltransferase sustains a core epigenetic programthat promotes metastatic colonization in breast cancer.',
'authors' => 'Couto J.P. et al.',
'description' => '<p><span>Metastatic colonization of distant organs accounts for over 90% of deaths related to solid cancers, yet the molecular determinants of metastasis remain poorly understood. Here, we unveil a mechanism of colonization in the aggressive basal-like subtype of breast cancer that is driven by the NAD</span><sup>+</sup><span><span> </span>metabolic enzyme nicotinamide N-methyltransferase (NNMT). We demonstrate that NNMT imprints a basal genetic program into cancer cells, enhancing their plasticity. In line, NNMT expression is associated with poor clinical outcomes in patients with breast cancer. Accordingly, ablation of NNMT dramatically suppresses metastasis formation in pre-clinical mouse models. Mechanistically, NNMT depletion results in a methyl overflow that increases histone H3K9 trimethylation (H3K9me3) and DNA methylation at the promoters of PR/SET Domain-5 (PRDM5) and extracellular matrix-related genes. PRDM5 emerged in this study as a pro-metastatic gene acting via induction of cancer-cell intrinsic transcription of collagens. Depletion of PRDM5 in tumor cells decreases COL1A1 deposition and impairs metastatic colonization of the lungs. These findings reveal a critical activity of the NNMT-PRDM5-COL1A1 axis for cancer cell plasticity and metastasis in basal-like breast cancer.</span></p>',
'date' => '2023-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37259596',
'doi' => '10.15252/embj.2022112559',
'modified' => '2023-06-15 08:35:19',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4787',
'name' => 'The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.',
'authors' => 'Garcia-Gomez E. et al.',
'description' => '<p>Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the disease (control group). In addition, eight of the PCOS patients underwent intervention with metformin (1500 mg/day) and a carbohydrate-controlled diet (type and quantity) for three months. Clinical and metabolic parameters were determined, and RT-qPCR and MeDIP-qPCR were used to evaluate gene expression and DNA methylation levels, respectively. Decreased expression levels of , , and genes and increased DNA methylation levels of the promoter were found in the endometrium of PCOS patients compared to controls. After metformin and nutritional intervention, some metabolic and clinical variables improved in PCOS patients. This intervention was associated with increased expression of , , and genes and reduced DNA methylation levels of the promoter in the endometrium of PCOS women. Our preliminary findings suggest that metformin and a carbohydrate-controlled diet improve endometrial function in PCOS patients, partly by modulating DNA methylation of the gene promoter and the expression of genes implicated in endometrial receptivity and insulin signaling.</p>',
'date' => '2023-04-01',
'pmid' => 'https://doi.org/10.3390%2Fijms24076857',
'doi' => '10.3390/ijms24076857',
'modified' => '2023-06-12 08:58:33',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4777',
'name' => 'Epigenetic modifier alpha-ketoglutarate modulates aberrant gene bodymethylation and hydroxymethylation marks in diabetic heart.',
'authors' => 'Dhat R. et al.',
'description' => '<p>BACKGROUND: Diabetic cardiomyopathy (DCM) is a leading cause of death in diabetic patients. Hyperglycemic myocardial microenvironment significantly alters chromatin architecture and the transcriptome, resulting in aberrant activation of signaling pathways in a diabetic heart. Epigenetic marks play vital roles in transcriptional reprogramming during the development of DCM. The current study is aimed to profile genome-wide DNA (hydroxy)methylation patterns in the hearts of control and streptozotocin (STZ)-induced diabetic rats and decipher the effect of modulation of DNA methylation by alpha-ketoglutarate (AKG), a TET enzyme cofactor, on the progression of DCM. METHODS: Diabetes was induced in male adult Wistar rats with an intraperitoneal injection of STZ. Diabetic and vehicle control animals were randomly divided into groups with/without AKG treatment. Cardiac function was monitored by performing cardiac catheterization. Global methylation (5mC) and hydroxymethylation (5hmC) patterns were mapped in the Left ventricular tissue of control and diabetic rats with the help of an enrichment-based (h)MEDIP-sequencing technique by using antibodies specific for 5mC and 5hmC. Sequencing data were validated by performing (h)MEDIP-qPCR analysis at the gene-specific level, and gene expression was analyzed by qPCR. The mRNA and protein expression of enzymes involved in the DNA methylation and demethylation cycle were analyzed by qPCR and western blotting. Global 5mC and 5hmC levels were also assessed in high glucose-treated DNMT3B knockdown H9c2 cells. RESULTS: We found the increased expression of DNMT3B, MBD2, and MeCP2 with a concomitant accumulation of 5mC and 5hmC, specifically in gene body regions of diabetic rat hearts compared to the control. Calcium signaling was the most significantly affected pathway by cytosine modifications in the diabetic heart. Additionally, hypermethylated gene body regions were associated with Rap1, apelin, and phosphatidyl inositol signaling, while metabolic pathways were most affected by hyperhydroxymethylation. AKG supplementation in diabetic rats reversed aberrant methylation patterns and restored cardiac function. Hyperglycemia also increased 5mC and 5hmC levels in H9c2 cells, which was normalized by DNMT3B knockdown or AKG supplementation. CONCLUSION: This study demonstrates that reverting hyperglycemic damage to cardiac tissue might be possible by erasing adverse epigenetic signatures by supplementing epigenetic modulators such as AKG along with an existing antidiabetic treatment regimen.</p>',
'date' => '2023-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37101286',
'doi' => '10.1186/s13072-023-00489-4',
'modified' => '2023-06-12 09:20:54',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '4780',
'name' => 'Integrated analysis from multicentre studies identities RNAmethylation- related lncRNA risk stratification systems for glioma',
'authors' => 'Huang Fanxuan and Wang Xinyu and Zhong Junzhe and Chen Hao and Song Dan and Xu Tianye and Tian Kaifu and Sun Penggang and Sun Nan and Ma Wenbin and Liu Yuxiang andYu Daohan and Meng Xiangqi and Jiang Chuanlu and Xuan Hanwen and Qian Da an',
'description' => '<p>Gastric cancer (GC) is the fourth leading cause of cancer death worldwide. Due to the lack of effective chemotherapy methods for advanced gastric cancer and poor prognosis, the emergence of immunotherapy has brought new hope to gastric cancer. Further research is needed to improve the response rate to immunotherapy and identify the populations with potential benefits of immunotherapy. It is unclear whether m7G-related lncRNAs influence tumour immunity and the prognosis of immunotherapy.</p>',
'date' => '2023-03-02',
'pmid' => 'https://doi.org/10.21203%2Frs.3.rs-2815231%2Fv1',
'doi' => '10.21203/rs.3.rs-2815231/v1',
'modified' => '2023-06-13 09:25:12',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '4773',
'name' => 'The RNA m5C Methylase NSUN2 Modulates Corneal EpithelialWound Healing.',
'authors' => 'Luo G. et al.',
'description' => '<p>PURPOSE: The emerging epitranscriptomics offers insights into the physiopathological roles of various RNA modifications. The RNA methylase NOP2/Sun domain family member 2 (NSUN2) catalyzes 5-methylcytosine (m5C) modification of mRNAs. However, the role of NSUN2 in corneal epithelial wound healing (CEWH) remains unknown. Here we describe the functional mechanisms of NSUN2 in mediating CEWH. METHODS: RT-qPCR, Western blot, dot blot, and ELISA were used to determine the NSUN2 expression and overall RNA m5C level during CEWH. NSUN2 silencing or overexpression was performed to explore its involvement in CEWH both in vivo and in vitro. Multi-omics was integrated to reveal the downstream target of NSUN2. MeRIP-qPCR, RIP-qPCR, and luciferase assay, as well as in vivo and in vitro functional assays, clarified the molecular mechanism of NSUN2 in CEWH. RESULTS: The NSUN2 expression and RNA m5C level increased significantly during CEWH. NSUN2 knockdown significantly delayed CEWH in vivo and inhibited human corneal epithelial cells (HCEC) proliferation and migration in vitro, whereas NSUN2 overexpression prominently enhanced HCEC proliferation and migration. Mechanistically, we found that NSUN2 increased ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) translation through the binding of RNA m5C reader Aly/REF export factor. Accordingly, UHRF1 knockdown significantly delayed CEWH in vivo and inhibited HCEC proliferation and migration in vitro. Furthermore, UHRF1 overexpression effectively rescued the inhibitory effect of NSUN2 silencing on HCEC proliferation and migration. CONCLUSIONS: NSUN2-mediated m5C modification of UHRF1 mRNA modulates CEWH. This finding highlights the critical importance of this novel epitranscriptomic mechanism in control of CEWH.</p>',
'date' => '2023-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36862118',
'doi' => '10.1167/iovs.64.3.5',
'modified' => '2023-04-17 09:48:55',
'created' => '2023-04-14 13:41:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '4674',
'name' => 'Methylation and expression of glucocorticoid receptor exon-1 variants andFKBP5 in teenage suicide-completers.',
'authors' => 'Rizavi H. et al.',
'description' => '<p>A dysregulated hypothalamic-pituitary-adrenal (HPA) axis has repeatedly been demonstrated to play a fundamental role in psychiatric disorders and suicide, yet the mechanisms underlying this dysregulation are not clear. Decreased expression of the glucocorticoid receptor (GR) gene, which is also susceptible to epigenetic modulation, is a strong indicator of impaired HPA axis control. In the context of teenage suicide-completers, we have systematically analyzed the 5'UTR of the GR gene to determine the expression levels of all GR exon-1 transcript variants and their epigenetic state. We also measured the expression and the epigenetic state of the FK506-binding protein 51 (FKBP5/FKBP51), an important modulator of GR activity. Furthermore, steady-state DNA methylation levels depend upon the interplay between enzymes that promote DNA methylation and demethylation activities, thus we analyzed DNA methyltransferases (DNMTs), ten-eleven translocation enzymes (TETs), and growth arrest- and DNA-damage-inducible proteins (GADD45). Focusing on both the prefrontal cortex (PFC) and hippocampus, our results show decreased expression in specific GR exon-1 variants and a strong correlation of DNA methylation changes with gene expression in the PFC. FKBP5 expression is also increased in both areas suggesting a decreased GR sensitivity to cortisol binding. We also identified aberrant expression of DNA methylating and demethylating enzymes in both brain regions. These findings enhance our understanding of the complex transcriptional regulation of GR, providing evidence of epigenetically mediated reprogramming of the GR gene, which could lead to possible epigenetic influences that result in lasting modifications underlying an individual's overall HPA axis response and resilience to stress.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36781843',
'doi' => '10.1038/s41398-023-02345-1',
'modified' => '2023-04-14 09:26:37',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '4675',
'name' => 'Bridging biological cfDNA features and machine learning approaches.',
'authors' => 'Moser T. et al.',
'description' => '<p>Liquid biopsies (LBs), particularly using circulating tumor DNA (ctDNA), are expected to revolutionize precision oncology and blood-based cancer screening. Recent technological improvements, in combination with the ever-growing understanding of cell-free DNA (cfDNA) biology, are enabling the detection of tumor-specific changes with extremely high resolution and new analysis concepts beyond genetic alterations, including methylomics, fragmentomics, and nucleosomics. The interrogation of a large number of markers and the high complexity of data render traditional correlation methods insufficient. In this regard, machine learning (ML) algorithms are increasingly being used to decipher disease- and tissue-specific signals from cfDNA. Here, we review recent insights into biological ctDNA features and how these are incorporated into sophisticated ML applications.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36792446',
'doi' => '10.1016/j.tig.2023.01.004',
'modified' => '2023-04-14 09:28:00',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '4823',
'name' => 'Gene body DNA hydroxymethylation restricts the magnitude oftranscriptional changes during aging.',
'authors' => 'Occean J. R. et al.',
'description' => '<p>DNA hydroxymethylation (5hmC) is the most abundant oxidative derivative of DNA methylation (5mC) and is typically enriched at enhancers and gene bodies of transcriptionally active and tissue-specific genes. Although aberrant genomic 5hmC has been implicated in many age-related diseases, the functional role of the modification in aging remains largely unknown. Here, we report that 5hmC is stably enriched in multiple aged organs. Using the liver and cerebellum as model organs, we show that 5hmC accumulates in gene bodies associated with tissue-specific function and thereby restricts the magnitude of gene expression changes during aging. Mechanistically, we found that 5hmC decreases binding affinity of splicing factors compared to unmodified cytosine and 5mC, and is correlated with age-related alternative splicing events, suggesting RNA splicing as a potential mediator of 5hmC’s transcriptionally restrictive function. Furthermore, we show that various age-related contexts, such as prolonged quiescence and senescence, are partially responsible for driving the accumulation of 5hmC with age. We provide evidence that this age-related function is conserved in mouse and human tissues, and further show that the modification is altered by regimens known to modulate lifespan. Our findings reveal that 5hmC is a regulator of tissue-specific function and may play a role in regulating longevity.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36824863',
'doi' => '10.1101/2023.02.15.528714',
'modified' => '2023-06-14 08:39:26',
'created' => '2023-06-13 22:16:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '4631',
'name' => 'Consistent DNA Hypomethylations in Prostate Cancer.',
'authors' => 'Araúzo-Bravo M.J. et al.',
'description' => '<p>With approximately 1.4 million men annually diagnosed with prostate cancer (PCa) worldwide, PCa remains a dreaded threat to life and source of devastating morbidity. In recent decades, a significant decrease in age-specific PCa mortality has been achieved by increasing prostate-specific antigen (PSA) screening and improving treatments. Nevertheless, upcoming, augmented recommendations against PSA screening underline an escalating disproportion between the benefit and harm of current diagnosis/prognosis and application of radical treatment standards. Undoubtedly, new potent diagnostic and prognostic tools are urgently needed to alleviate this tensed situation. They should allow a more reliable early assessment of the upcoming threat, in order to enable applying timely adjusted and personalized therapy and monitoring. Here, we present a basic study on an epigenetic screening approach by Methylated DNA Immunoprecipitation (MeDIP). We identified genes associated with hypomethylated CpG islands in three PCa sample cohorts. By adjusting our computational biology analyses to focus on single CpG-enriched 60-nucleotide-long DNA probes, we revealed numerous consistently differential methylated DNA segments in PCa. They were associated among other genes with and . These can be used for early discrimination, and might contribute to a new epigenetic tumor classification system of PCa. Our analysis shows that we can dissect short, differential methylated CpG-rich DNA fragments and combinations of them that are consistently present in all tumors. We name them tumor cell-specific differential methylated CpG dinucleotide signatures (TUMS).</p>',
'date' => '2022-12-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36613831',
'doi' => '10.3390/ijms24010386',
'modified' => '2023-03-28 09:03:47',
'created' => '2023-02-21 09:59:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '4534',
'name' => 'RNA 5-Methylcytosine Modification Regulates VegetativeDevelopment Associated with H3K27 Trimethylation inArabidopsis.',
'authors' => 'Zhang D.et al.',
'description' => '<p>Methylating RNA post-transcriptionally is emerging as a significant mechanism of gene regulation in eukaryotes. The crosstalk between RNA methylation and histone modification is critical for chromatin state and gene expression in mammals. However, it is not well understood mechanistically in plants. Here, the authors report a genome-wide correlation between RNA 5-cytosine methylation (m C) and histone 3 lysine27 trimethylation (H3K27me3) in Arabidopsis. The plant-specific Polycomb group (PcG) protein EMBRYONIC FLOWER1 (EMF1) plays dual roles as activators or repressors. Transcriptome-wide RNA m C profiling revealed that m C peaks are mostly enriched in chromatin regions that lacked H3K27me3 in both wild type and emf1 mutants. EMF1 repressed the expression of m C methyltransferase tRNA specific methyltransferase 4B (TRM4B) through H3K4me3, independent of PcG-mediated H3K27me3 mechanism. The 5-Cytosine methylation on targets is increased in emf1 mutants, thereby decreased the mRNA transcripts of photosynthesis and chloroplast genes. In addition, impairing EMF1 activity reduced H3K27me3 levels of PcG targets, such as starch genes, which are de-repressed in emf1 mutants. Both EMF1-mediated promotion and repression of gene activities via m C and H3K27me3 are required for normal vegetative growth. Collectively, t study reveals a previously undescribed epigenetic mechanism of RNA m C modifications and histone modifications to regulate gene expression in eukaryotes.</p>',
'date' => '2022-11-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36382558',
'doi' => '10.1002/advs.202204885',
'modified' => '2022-11-24 08:57:31',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '4541',
'name' => 'Cardiac epigenetic changes in VEGF signaling genes associates with myocardial microvascular rarefaction in experimental chronic kidney disease.',
'authors' => 'Eirin Alfonso and Chade Alejandro R',
'description' => '<p>BACKGROUND: Chronic kidney disease (CKD) is common in patients with heart failure, and often results in left ventricular diastolic dysfunction (LVDD). However, the mechanisms responsible for cardiac damage in CKD-LVDD remain to be elucidated. Epigenetic alterations may impose long-lasting effects on cellular transcription and function, but their exact role in CKD-LVDD is unknown. We investigate whether changes in cardiac site-specific DNA methylation profiles might be implicated in cardiac abnormalities in CKD-LVDD. METHODS: CKD-LVDD and normal control pigs (n=6 each) were studied for 14 weeks. Renal and cardiac hemodynamics were quantified by multidetector CT and echocardiography. In randomly selected pigs (n=3/group), cardiac site-specific 5-methylcytosine (5mC) immunoprecipitation (MeDIP)- and mRNA-sequencing (seq) was performed, followed by integrated (MeDiP-seq/mRNA-seq analysis), and confirmatory ex vivo studies. RESULTS: MeDIP-seq analysis revealed 261 genes with higher (fold-change>1.4; p<0.05) and 162 genes with lower (fold-change<0.7; p<0.05) 5mC levels in CKD-LVDD versus normal pigs, which were primarily implicated in vascular endothelial growth factor (VEGF)-related signaling and angiogenesis. Integrated MeDiP-seq/mRNA-seq analysis identified a select group of VEGF-related genes in which 5mC levels were higher, but mRNA expression lower in CKD-LVDD versus normal pigs. Cardiac VEGF signaling gene and VEGF protein expression was blunted in CKD-LVDD compared to controls and associated with decreased subendocardial microvascular density. CONCLUSIONS: Cardiac epigenetic changes in VEGF-related genes are associated with impaired angiogenesis and cardiac microvascular rarefaction in swine CKD-LVDD. These observations may assist in developing novel therapies to ameliorate cardiac damage in CKD-LVDD.</p>',
'date' => '2022-11-01',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/36367693/',
'doi' => '10.1152/ajpheart.00522.2022',
'modified' => '2022-11-25 09:03:31',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '4511',
'name' => 'The Arabidopsis APOLO and human UPAT sequence-unrelated longnoncoding RNAs can modulate DNA and histone methylation machineries inplants.',
'authors' => 'Fonouni-Farde C. et al.',
'description' => '<p>BACKGROUND: RNA-DNA hybrid (R-loop)-associated long noncoding RNAs (lncRNAs), including the Arabidopsis lncRNA AUXIN-REGULATED PROMOTER LOOP (APOLO), are emerging as important regulators of three-dimensional chromatin conformation and gene transcriptional activity. RESULTS: Here, we show that in addition to the PRC1-component LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), APOLO interacts with the methylcytosine-binding protein VARIANT IN METHYLATION 1 (VIM1), a conserved homolog of the mammalian DNA methylation regulator UBIQUITIN-LIKE CONTAINING PHD AND RING FINGER DOMAINS 1 (UHRF1). The APOLO-VIM1-LHP1 complex directly regulates the transcription of the auxin biosynthesis gene YUCCA2 by dynamically determining DNA methylation and H3K27me3 deposition over its promoter during the plant thermomorphogenic response. Strikingly, we demonstrate that the lncRNA UHRF1 Protein Associated Transcript (UPAT), a direct interactor of UHRF1 in humans, can be recognized by VIM1 and LHP1 in plant cells, despite the lack of sequence homology between UPAT and APOLO. In addition, we show that increased levels of APOLO or UPAT hamper VIM1 and LHP1 binding to YUCCA2 promoter and globally alter the Arabidopsis transcriptome in a similar manner. CONCLUSIONS: Collectively, our results uncover a new mechanism in which a plant lncRNA coordinates Polycomb action and DNA methylation through the interaction with VIM1, and indicates that evolutionary unrelated lncRNAs with potentially conserved structures may exert similar functions by interacting with homolog partners.</p>',
'date' => '2022-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36038910',
'doi' => '10.1186/s13059-022-02750-7',
'modified' => '2022-11-21 10:43:16',
'created' => '2022-11-15 09:26:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '4438',
'name' => 'A genome-wide screen reveals new regulators of the 2-cell-like cell state',
'authors' => 'Defossez Pierre-Antoine et al.',
'description' => '<p>In mammals, only the zygote and blastomeres of the early embryo are fully totipotent. This totipotency is mirrored in vitro by mouse "2-cell-like cells" (2CLCs), which appear at low frequency in cultures of Embryonic Stem cells (ESCs). Because totipotency is incompletely understood, we carried out a genomewide CRISPR KO screen in mouse ESCs, searching for mutants that reactivate the expression of Dazl, a robust 2-cell-like marker. Using secondary screens, we identify four mutants that reactivate not just Dazl, but also a broader 2-cell-like signature: the E3 ubiquitin ligase adaptor SPOP, the Zinc Finger transcription factor ZBTB14, MCM3AP, a component of the RNA processing complex TREX-2, and the lysine demethylase KDM5C. Functional experiments show how these factors link to known players of the 2 celllike state. These results extend our knowledge of totipotency, a key phase of organismal life.</p>',
'date' => '2022-06-01',
'pmid' => 'https://doi.org/10.21203%2Frs.3.rs-1561018%2Fv1',
'doi' => '10.21203/rs.3.rs-1561018/v1',
'modified' => '2022-09-28 09:23:42',
'created' => '2022-09-08 16:32:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '4553',
'name' => 'NSUN2-mediated RNA mC modification modulates uveal melanoma cellproliferation and migration.',
'authors' => 'Luo Guangying et al.',
'description' => '<p>RNA 5-methylcytosine (mC) is a widespread post-transcriptional modification involved in diverse biological processes through controlling RNA metabolism. However, its roles in uveal melanoma (UM) remain unknown. Here, we describe the biological roles and regulatory mechanisms of RNA mC in UM. Initially, we identified significantly elevated global RNA mC levels in both UM cells and tissue specimens using ELISA assay and dot blot analysis. Meanwhile, NOP2/Sun RNA methyltransferase family member 2 (NSUN2) was upregulated in both types of these samples, whereas NSUN2 knockdown significantly decreased RNA mC level. Such declines inhibited UM cell migration and suppressed cell proliferation through cell cycle G1 arrest. Furthermore, bioinformatic analyses, mC-RIP-qPCR, and luciferase assay identified β-Catenin (CTNNB1) as a direct target of NSUN2-mediated mC modification in UM cells. Additionally, overexpression of miR-124a in UM cells diminished NSUN2 expression levels indicating that it is an upstream regulator of this response. Our study suggests that NSUN2-mediated RNA mC methylation provides a potential novel target to improve the therapeutic management of UM pathogenesis.</p>',
'date' => '2022-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/35757999',
'doi' => '10.1080/15592294.2022.2088047',
'modified' => '2022-11-24 10:14:24',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '4340',
'name' => 'Global DNA methylation and cellular 5-methylcytosine and H4acetylated patterns in primary and secondary dormant seeds of Capsellabursa-pastoris (L.) Medik. (shepherd's purse).',
'authors' => 'Gomez-Cabellos Sara et al.',
'description' => '<p>Despite the importance of dormancy and dormancy cycling for plants' fitness and life cycle phenology, a comprehensive characterization of the global and cellular epigenetic patterns across space and time in different seed dormancy states is lacking. Using Capsella bursa-pastoris (L.) Medik. (shepherd's purse) seeds with primary and secondary dormancy, we investigated the dynamics of global genomic DNA methylation and explored the spatio-temporal distribution of 5-methylcytosine (5-mC) and histone H4 acetylated (H4Ac) epigenetic marks. Seeds were imbibed at 30 °C in a light regime to maintain primary dormancy, or in darkness to induce secondary dormancy. An ELISA-based method was used to quantify DNA methylation, in relation to total genomic cytosines. Immunolocalization of 5-mC and H4Ac within whole seeds (i.e., including testa) was assessed with reference to embryo anatomy. Global DNA methylation levels were highest in prolonged (14 days) imbibed primary dormant seeds, with more 5-mC marked nuclei present only in specific parts of the seed (e.g., SAM and cotyledons). In secondary dormant seeds, global methylation levels and 5-mC signal where higher at 3 and 7 days than 1 or 14 days. With respect to acetylation, seeds had fewer H4Ac marked nuclei (e.g., SAM) in deeper dormant states, for both types of dormancy. However, the RAM still showed signal after 14 days of imbibition under dormancy-inducing conditions, suggesting a central role for the radicle/RAM in the response to perceived ambient changes and the adjustment of the seed dormancy state. Thus, we show that seed dormancy involves extensive cellular remodeling of DNA methylation and H4 acetylation.</p>',
'date' => '2022-05-01',
'pmid' => 'https://doi.org/10.1007%2Fs00709-021-01678-2',
'doi' => '10.1007/s00709-021-01678-2',
'modified' => '2022-06-20 09:19:49',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '4404',
'name' => 'Stella regulates the Development of Female Germline Stem Cells byModulating Chromatin Structure and DNA Methylation.',
'authors' => 'Hou Changliang et al.',
'description' => '<p>Female germline stem cells (FGSCs) have the ability to self-renew and differentiate into oocytes. , encoded by a maternal effect gene, plays an important role in oogenesis and early embryonic development. However, its function in FGSCs remains unclear. In this study, we showed that CRISPR/Cas9-mediated knockout of promoted FGSC proliferation and reduced the level of genome-wide DNA methylation of FGSCs. Conversely, overexpression led to the opposite results, and enhanced FGSC differentiation. We also performed an integrative analysis of chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), high-throughput genome-wide chromosome conformation capture (Hi-C), and use of our published epigenetic data. Results indicated that the binding sites of STELLA and active histones H3K4me3 and H3K27ac were enriched near the TAD boundaries. Hi-C analysis showed that overexpression attenuated the interaction within TADs, and interestingly enhanced the TAD boundary strength in STELLA-associated regions. Taking these findings together, our study not only reveals the role of in regulating DNA methylation and chromatin structure, but also provides a better understanding of FGSC development.</p>',
'date' => '2022-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9066111/',
'doi' => '10.7150/ijbs.69240',
'modified' => '2022-08-11 14:54:29',
'created' => '2022-08-11 12:14:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '4327',
'name' => 'Highly recurrent epimutations in gastric cancer CpG islandmethylator phenotypes and inflammation',
'authors' => 'Padmanabhan N. et al.',
'description' => '<p>Background CIMP (CpG island methylator phenotype) is an epigenetic molecular subtype, observed in multiple malignancies and associated with the epigenetic silencing of tumor suppressors. Currently, for most cancers including gastric cancer (GC), mechanisms underlying CIMP remain poorly understood. We sought to discover molecular contributors to CIMP in GC, by performing global DNA methylation, gene expression, and proteomics profiling across 14 gastric cell lines, followed by similar integrative analysis in 50 GC cell lines and 467 primary GCs. Results We identify the cystathionine beta-synthase enzyme (CBS) as a highly recurrent target of epigenetic silencing in CIMP GC. Likewise, we show that CBS epimutations are significantly associated with CIMP in various other cancers, occurring even in premalignant gastroesophageal conditions and longitudinally linked to clinical persistence. Of note, CRISPR deletion of CBS in normal gastric epithelial cells induces widespread DNA methylation changes that overlap with primary GC CIMP patterns. Reflecting its metabolic role as a gatekeeper interlinking the methionine and homocysteine cycles, CBS loss in vitro also causes reductions in the anti-inflammatory gasotransmitter hydrogen sulfide (H2S), with concomitant increase in NF-κB activity. In a murine genetic model of CBS deficiency, preliminary data indicate upregulated immune-mediated transcriptional signatures in the stomach. Conclusions Our results implicate CBS as a bi-faceted modifier of aberrant DNA methylation and inflammation in GC and highlights H2S donors as a potential new therapy for CBS-silenced lesions. Supplementary Information The online version contains supplementary material available at 10.1186/s13059-021-02375-2.</p>',
'date' => '2021-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/34074348',
'doi' => '10.1186/s13059-021-02375-2',
'modified' => '2022-08-03 16:01:40',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '4336',
'name' => 'LINE-1 transcription in round spermatids is associated with accretion of5-carboxylcytosine in their open reading frames',
'authors' => 'Blythe M. et al. ',
'description' => '<p>Chromatin of male and female gametes undergoes a number of reprogramming events during the transition from germ cell to embryonic developmental programs. Although the rearrangement of DNA methylation patterns occurring in the zygote has been extensively characterized, little is known about the dynamics of DNA modifications during spermatid maturation. Here, we demonstrate that the dynamics of 5-carboxylcytosine (5caC) correlate with active transcription of LINE-1 retroelements during murine spermiogenesis. We show that the open reading frames of active and evolutionary young LINE-1s are 5caC-enriched in round spermatids and 5caC is eliminated from LINE-1s and spermiogenesis-specific genes during spermatid maturation, being simultaneously retained at promoters and introns of developmental genes. Our results reveal an association of 5caC with activity of LINE-1 retrotransposons suggesting a potential direct role for this DNA modification in fine regulation of their transcription.</p>',
'date' => '2021-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/34099857',
'doi' => '10.1038/s42003-021-02217-8',
'modified' => '2022-08-03 16:17:04',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '4150',
'name' => 'Sensitive and reproducible cell-free methylome quantification with synthetic spike-in controls',
'authors' => 'Wilson, S.L. et al.',
'description' => '<p>Background. Cell-free methylated DNA immunoprecipitation-sequencing (cfMeDIP-seq) identifies genomic regions with DNA methylation, using a protocol adapted to work with low-input DNA samples and with cell-free DNA (cfDNA). This method allows for DNA methylation profiling of circulating tumour DNA in cancer patients’ blood samples. Such epigenetic profiling of circulating tumour DNA provides information about in which tissues tumour DNA originates, a key requirement of any test for early cancer detection. In addition, DNA methylation signatures provide prognostic information and can detect relapse. For robust quantitative comparisons between samples, immunoprecipitation enrichment methods like cfMeDIP-seq require normalization against common reference controls.</p>',
'date' => '2021-04-01',
'pmid' => 'https://doi.org/10.1101%2F2021.02.12.430289',
'doi' => '10.1101/2021.02.12.430289',
'modified' => '2022-01-13 15:16:23',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '3985',
'name' => 'Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes.',
'authors' => 'Nuzzo PV, Berchuck JE, Korthauer K, Spisak S, Nassar AH, Abou Alaiwi S, Chakravarthy A, Shen SY, Bakouny Z, Boccardo F, Steinharter J, Bouchard G, Curran CR, Pan W, Baca SC, Seo JH, Lee GM, Michaelson MD, Chang SL, Waikar SS, Sonpavde G, Irizarry RA, Pome',
'description' => '<p>Improving early cancer detection has the potential to substantially reduce cancer-related mortality. Cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) is a highly sensitive assay capable of detecting early-stage tumors. We report accurate classification of patients across all stages of renal cell carcinoma (RCC) in plasma (area under the receiver operating characteristic (AUROC) curve of 0.99) and demonstrate the validity of this assay to identify patients with RCC using urine cell-free DNA (cfDNA; AUROC of 0.86).</p>',
'date' => '2020-06-22',
'pmid' => 'http://www.pubmed.gov/32572266',
'doi' => '10.1038/s41591-020-0933-1',
'modified' => '2020-09-01 15:13:49',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '3984',
'name' => 'Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes.',
'authors' => 'Nassiri F, Chakravarthy A, Feng S, Shen SY, Nejad R, Zuccato JA, Voisin MR, Patil V, Horbinski C, Aldape K, Zadeh G, De Carvalho DD',
'description' => '<p>Definitive diagnosis of intracranial tumors relies on tissue specimens obtained by invasive surgery. Noninvasive diagnostic approaches provide an opportunity to avoid surgery and mitigate unnecessary risk to patients. In the present study, we show that DNA-methylation profiles from plasma reveal highly specific signatures to detect and accurately discriminate common primary intracranial tumors that share cell-of-origin lineages and can be challenging to distinguish using standard-of-care imaging.</p>',
'date' => '2020-06-22',
'pmid' => 'http://www.pubmed.gov/32572265',
'doi' => '10.1038/s41591-020-0932-2',
'modified' => '2020-09-01 15:14:45',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '4030',
'name' => 'AXR1 affects DNA methylation independently of its role in regulatingmeiotic crossover localization.',
'authors' => 'Christophorou, N and She, W and Long, J and Hurel, A and Beaubiat, S andIdir, Y and Tagliaro-Jahns, M and Chambon, A and Solier, V and Vezon, D andGrelon, M and Feng, X and Bouché, N and Mézard, C',
'description' => '<p>Meiotic crossovers (COs) are important for reshuffling genetic information between homologous chromosomes and they are essential for their correct segregation. COs are unevenly distributed along chromosomes and the underlying mechanisms controlling CO localization are not well understood. We previously showed that meiotic COs are mis-localized in the absence of AXR1, an enzyme involved in the neddylation/rubylation protein modification pathway in Arabidopsis thaliana. Here, we report that in axr1-/-, male meiocytes show a strong defect in chromosome pairing whereas the formation of the telomere bouquet is not affected. COs are also redistributed towards subtelomeric chromosomal ends where they frequently form clusters, in contrast to large central regions depleted in recombination. The CO suppressed regions correlate with DNA hypermethylation of transposable elements (TEs) in the CHH context in axr1-/- meiocytes. Through examining somatic methylomes, we found axr1-/- affects DNA methylation in a plant, causing hypermethylation in all sequence contexts (CG, CHG and CHH) in TEs. Impairment of the main pathways involved in DNA methylation is epistatic over axr1-/- for DNA methylation in somatic cells but does not restore regular chromosome segregation during meiosis. Collectively, our findings reveal that the neddylation pathway not only regulates hormonal perception and CO distribution but is also, directly or indirectly, a major limiting pathway of TE DNA methylation in somatic cells.</p>',
'date' => '2020-06-01',
'pmid' => 'http://www.pubmed.gov/32598340',
'doi' => '10.1371/journal.',
'modified' => '2020-12-16 17:58:51',
'created' => '2020-10-12 14:54:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '3951',
'name' => 'In vitro capture and characterization of embryonic rosette-stage pluripotency between naive and primed states.',
'authors' => 'Neagu A, van Genderen E, Escudero I, Verwegen L, Kurek D, Lehmann J, Stel J, Dirks RAM, van Mierlo G, Maas A, Eleveld C, Ge Y, den Dekker AT, Brouwer RWW, van IJcken WFJ, Modic M, Drukker M, Jansen JH, Rivron NC, Baart EB, Marks H, Ten Berge D',
'description' => '<p>Following implantation, the naive pluripotent epiblast of the mouse blastocyst generates a rosette, undergoes lumenogenesis and forms the primed pluripotent egg cylinder, which is able to generate the embryonic tissues. How pluripotency progression and morphogenesis are linked and whether intermediate pluripotent states exist remain controversial. We identify here a rosette pluripotent state defined by the co-expression of naive factors with the transcription factor OTX2. Downregulation of blastocyst WNT signals drives the transition into rosette pluripotency by inducing OTX2. The rosette then activates MEK signals that induce lumenogenesis and drive progression to primed pluripotency. Consequently, combined WNT and MEK inhibition supports rosette-like stem cells, a self-renewing naive-primed intermediate. Rosette-like stem cells erase constitutive heterochromatin marks and display a primed chromatin landscape, with bivalently marked primed pluripotency genes. Nonetheless, WNT induces reversion to naive pluripotency. The rosette is therefore a reversible pluripotent intermediate whereby control over both pluripotency progression and morphogenesis pivots from WNT to MEK signals.</p>',
'date' => '2020-05-01',
'pmid' => 'http://www.pubmed.gov/32367046',
'doi' => '10.1038/s41556-020-0508-x',
'modified' => '2020-08-17 09:55:37',
'created' => '2020-08-10 12:12:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '3834',
'name' => 'Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.',
'authors' => 'Liu J, An Z, Luo J, Li J, Li F, Zhang Z',
'description' => '<p>MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of transcriptome-wide profile and potential function of m5C in splicing remains to be elucidated due to lack of isoform level m5C quantification tool. RESULTS: We developed a computational package to quantify Epitranscriptomal RNA m5C at the transcript isoform level (named Episo). Episo consists of three tools, mapper, quant and Bisulfitefq, for mapping, quantifying, and simulating RNA-BisSeq data, respectively. The high accuracy of Episo was validated using an improved m5C-specific methylated RNA immunoprecipitation (meRIP) protocol, as well as a set of in silico experiments. By applying Episo to public human and mouse RNA-BisSeq data, we found that the RNA m5C is not evenly distributed among the transcript isoforms, implying the m5C may subject to be regulated at isoform level. AVAILABILITY: Episo is released under the GNU GPLv3+ license. The resource code Episo is freely accessible from https://github.com/liujunfengtop/Episo (with Tophat/cufflink) and https://github.com/liujunfengtop/Episo/tree/master/Episo_Kallisto (with Kallisto). SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.</p>',
'date' => '2019-12-03',
'pmid' => 'http://www.pubmed.gov/31794005',
'doi' => '10.1093/bioinformatics/btz900/5651015',
'modified' => '2020-02-25 13:26:22',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 25 => array(
'id' => '3827',
'name' => 'Intra- and inter-generational changes in the cortical DNA methylome in response to therapeutic intermittent hypoxia in mice.',
'authors' => 'Belmonte KCD, Harman JC, Lanson NA, Gidday JM',
'description' => '<p>Recent evidence from our lab documents functional resilience to retinal ischemic injury in untreated mice derived from parents exposed to repetitive hypoxic conditioning (RHC) prior to breeding. To begin to understand the epigenetic basis of this intergenerational protection, we used methylated DNA immunoprecipitation and sequencing to identify genes with differentially-methylated promoters (DMGPs) in the prefrontal cortex of mice treated directly with the same RHC stimulus (F0-RHC), and in the prefrontal cortex of their untreated F1-generation offspring (F1-*RHC). Subsequent bioinformatic analyses provided key mechanistic insights into how changes in gene expression secondary to promoter hypo- and hyper-methylation might afford such protection within and across generations. We found extensive changes in DNA methylation in both generations consistent with the expression of many survival-promoting genes, with twice the number of DMGPs in the cortex of F1*RHC mice relative to their F0 parents that were directly exposed to RHC. In contrast to our hypothesis that similar epigenetic modifications would be realized in the cortices of both F0-RHC and F1-*RHC mice, we instead found relatively few DMGPs common to both generations; in fact, each generation manifested expected injury resilience via distinctly unique gene expression profiles. Whereas in the cortex of F0-RHC mice, predicted protein-protein interactions reflected the activation of an anti-ischemic phenotype, networks activated in F1-*RHC cortex comprised networks indicative of a much broader cytoprotective phenotype. Altogether, our results suggest that the intergenerational transfer of an acquired phenotype to offspring does not necessarily require the faithful recapitulation of the conditioning-modified DNA methylome of the parent.</p>',
'date' => '2019-11-25',
'pmid' => 'http://www.pubmed.gov/31762411',
'doi' => '10.1152/physiolgenomics.00094.2019',
'modified' => '2020-02-25 13:35:09',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 26 => array(
'id' => '3814',
'name' => 'Lithium treatment reverses irradiation-induced changes in rodent neural progenitors and rescues cognition.',
'authors' => 'Zanni G, Goto S, Fragopoulou AF, Gaudenzi G, Naidoo V, Di Martino E, Levy G, Dominguez CA, Dethlefsen O, Cedazo-Minguez A, Merino-Serrais P, Stamatakis A, Hermanson O, Blomgren K',
'description' => '<p>Cranial radiotherapy in children has detrimental effects on cognition, mood, and social competence in young cancer survivors. Treatments harnessing hippocampal neurogenesis are currently of great relevance in this context. Lithium, a well-known mood stabilizer, has both neuroprotective, pro-neurogenic as well as antitumor effects, and in the current study we introduced lithium treatment 4 weeks after irradiation. Female mice received a single 4 Gy whole-brain radiation dose on postnatal day (PND) 21 and were randomized to 0.24% Li2CO chow or normal chow from PND 49 to 77. Hippocampal neurogenesis was assessed on PND 77, 91, and 105. We found that lithium treatment had a pro-proliferative effect on neural progenitors, but neuronal integration occurred only after it was discontinued. Also, the treatment ameliorated deficits in spatial learning and memory retention observed in irradiated mice. Gene expression profiling and DNA methylation analysis identified two novel factors related to the observed effects, Tppp, associated with microtubule stabilization, and GAD2/65, associated with neuronal signaling. Our results show that lithium treatment reverses irradiation-induced loss of hippocampal neurogenesis and cognitive impairment even when introduced long after the injury. We propose that lithium treatment should be intermittent in order to first make neural progenitors proliferate and then, upon discontinuation, allow them to differentiate. Our findings suggest that pharmacological treatment of cognitive so-called late effects in childhood cancer survivors is possible.</p>',
'date' => '2019-11-14',
'pmid' => 'http://www.pubmed.gov/31723242',
'doi' => '10.1038/s41380-019-0584-0',
'modified' => '2019-12-05 10:58:44',
'created' => '2019-12-02 15:25:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 27 => array(
'id' => '3773',
'name' => 'Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA.',
'authors' => 'Shen SY, Burgener JM, Bratman SV, De Carvalho DD',
'description' => '<p>Circulating cell-free DNA (cfDNA) comprises small DNA fragments derived from normal and tumor tissue that are released into the bloodstream. Recently, methylation profiling of cfDNA as a liquid biopsy tool has been gaining prominence due to the presence of tissue-specific markers in cfDNA. We have previously reported cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) as a sensitive, low-input, cost-efficient and bisulfite-free approach to profiling DNA methylomes of plasma cfDNA. cfMeDIP-seq is an extension of a previously published MeDIP-seq protocol and is adapted to allow for methylome profiling of samples with low input (ranging from 1 to 10 ng) of DNA, which is enabled by the addition of 'filler DNA' before immunoprecipitation. This protocol is not limited to plasma cfDNA; it can also be applied to other samples that are naturally sheared and at low availability (e.g., urinary cfDNA and cerebrospinal fluid cfDNA), and is potentially applicable to other applications beyond cancer detection, including prenatal diagnostics, cardiology and monitoring of immune response. The protocol presented here should enable any standard molecular laboratory to generate cfMeDIP-seq libraries from plasma cfDNA in ~3-4 d.</p>',
'date' => '2019-08-30',
'pmid' => 'http://www.pubmed.gov/31471598',
'doi' => '10.1038/s41596-019-0202-2',
'modified' => '2019-10-02 17:07:45',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 28 => array(
'id' => '3763',
'name' => 'Silencing of tumor-suppressive NR_023387 in renal cell carcinoma via promoter hypermethylation and HNF4A deficiency.',
'authors' => 'Zhou H, Guo L, Yao W, Shi R, Yu G, Xu H, Ye Z',
'description' => '<p>Dysregulation of the epigenetic status of long noncoding RNAs (lncRNAs) has been linked to diverse human diseases including human cancers. However, the landscape of the whole-genome methylation profile of lncRNAs and the precise roles of these lncRNAs remain elusive in renal cell carcinoma (RCC). We first examined lncRNA expression profiles in RCC tissues and corresponding adjacent normal tissues (NTs) to identify the lncRNA signature of RCC, then lncRNA Promoter Microarray was performed to depict the whole-genome methylation profile of lncRNAs in RCC. Combined analysis of the lncRNAs expression profiles and lncRNAs Promoter Microarray identified a series of downregulated lncRNAs with hypermethylated promoter regions, including NR_023387. Quantitative real-time polymerase chain reaction (RT-PCR) implied that NR_023387 was significantly downregulated in RCC tissues and cell lines, and lower expression of NR_023387 was correlated with shorter overall survival. Methylation-specific PCR, MassARRAY, and demethylation drug treatment indicated that hypermethylation in the NR_023387 promoter contributed to its silencing in RCC. Besides, HNF4A regulated the expression of NR_023387 via transcriptional activation. Functional experiments demonstrated NR_023387 exerted tumor-suppressive roles in RCC via suppressing the proliferation, migration, invasion, tumor growth, and metastasis of RCC. Furthermore, we identified MGP as a putative downstream molecule of NR_023387, which promoted the epithelial-mesenchymal transition of RCC cells. Our study provides the first whole-genome lncRNA methylation profile in RCC. Our combined analysis identifies a tumor-suppressive and prognosis-related lncRNA NR_023387, which is silenced in RCC via promoter hypermethylation and HNF4A deficiency, and may exert its tumor-suppressive roles by downregulating the oncogenic MGP.</p>',
'date' => '2019-08-20',
'pmid' => 'http://www.pubmed.gov/31432508',
'doi' => '10.1002/jcp.29115',
'modified' => '2019-10-03 10:02:27',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 29 => array(
'id' => '3770',
'name' => 'Epitranscriptomic Addition of mC to HIV-1 Transcripts Regulates Viral Gene Expression.',
'authors' => 'Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR',
'description' => '<p>How the covalent modification of mRNA ribonucleotides, termed epitranscriptomic modifications, alters mRNA function remains unclear. One issue has been the difficulty of quantifying these modifications. Using purified HIV-1 genomic RNA, we show that this RNA bears more epitranscriptomic modifications than the average cellular mRNA, with 5-methylcytosine (mC) and 2'O-methyl modifications being particularly prevalent. The methyltransferase NSUN2 serves as the primary writer for mC on HIV-1 RNAs. NSUN2 inactivation inhibits not only mC addition to HIV-1 transcripts but also viral replication. This inhibition results from reduced HIV-1 protein, but not mRNA, expression, which in turn correlates with reduced ribosome binding to viral mRNAs. In addition, loss of mC dysregulates the alternative splicing of viral RNAs. These data identify mC as a post-transcriptional regulator of both splicing and function of HIV-1 mRNA, thereby affecting directly viral gene expression.</p>',
'date' => '2019-08-14',
'pmid' => 'http://www.pubmed.gov/31415754',
'doi' => '10.1016/j.chom.2019.07.005',
'modified' => '2019-10-03 09:18:50',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 30 => array(
'id' => '3741',
'name' => 'Aberrant expression of imprinted lncRNA MEG8 causes trophoblast dysfunction and abortion.',
'authors' => 'Sheng F, Sun N, Ji Y, Ma Y, Ding H, Zhang Q, Yang F, Li W',
'description' => '<p>Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs whose nucleotides are longer than 200 bp. Previous studies have shown that they play an important regulatory role in many developmental processes and biological pathways. However, the contributions of lncRNAs to placental development are largely unknown. Here, our study aimed to investigate the lncRNA expression signatures in placental development by performing a microarray lncRNA screen. Placental samples were obtained from pregnant C57BL/6 female mice at three key developmental time points (embryonic day E7.5, E13.5, and E19.5). Microarrays were used to analyze the differential expression of lncRNAs during placental development. In addition to the genomic imprinting region and the dynamic DNA methylation status during placental development, we screened imprinted lncRNAs whose expression was controlled by DNA methylation during placental development. We found that the imprinted lncRNA Rian may play an important role during placental development. Its homologous sequence lncRNA MEG8 (RIAN) was abnormally highly expressed in human spontaneous abortion villi. Upregulation of MEG8 expression in trophoblast cell lines decreased cell proliferation and invasion, whereas downregulation of MEG8 expression had the opposite effect. Furthermore, DNA methylation results showed that the methylation of the MEG8 promoter region was increased in spontaneous abortion villi. There was dynamic spatiotemporal expression of imprinted lncRNAs during placental development. The imprinted lncRNA MEG8 is involved in the regulation of early trophoblast cell function. Promoter methylation abnormalities can cause trophoblastic cell defects, which may be one of the factors that occurs in early unexplained spontaneous abortion.</p>',
'date' => '2019-07-02',
'pmid' => 'http://www.pubmed.gov/31265183',
'doi' => '10.1002/jcb.29002',
'modified' => '2019-08-06 16:45:53',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 31 => array(
'id' => '3731',
'name' => 'Defining UHRF1 Domains that Support Maintenance of Human Colon Cancer DNA Methylation and Oncogenic Properties.',
'authors' => 'Kong X, Chen J, Xie W, Brown SM, Cai Y, Wu K, Fan D, Nie Y, Yegnasubramanian S, Tiedemann RL, Tao Y, Chiu Yen RW, Topper MJ, Zahnow CA, Easwaran H, Rothbart SB, Xia L, Baylin SB',
'description' => '<p>UHRF1 facilitates the establishment and maintenance of DNA methylation patterns in mammalian cells. The establishment domains are defined, including E3 ligase function, but the maintenance domains are poorly characterized. Here, we demonstrate that UHRF1 histone- and hemimethylated DNA binding functions, but not E3 ligase activity, maintain cancer-specific DNA methylation in human colorectal cancer (CRC) cells. Disrupting either chromatin reader activity reverses DNA hypermethylation, reactivates epigenetically silenced tumor suppressor genes (TSGs), and reduces CRC oncogenic properties. Moreover, an inverse correlation between high UHRF1 and low TSG expression tracks with CRC progression and reduced patient survival. Defining critical UHRF1 domain functions and its relationship with CRC prognosis suggests directions for, and value of, targeting this protein to develop therapeutic DNA demethylating agents.</p>',
'date' => '2019-04-15',
'pmid' => 'http://www.pubmed.gov/30956060',
'doi' => '10.1016/j.ccell.2019.03.003',
'modified' => '2019-08-07 09:14:54',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 32 => array(
'id' => '3729',
'name' => 'Tricarboxylic Acid Cycle Activity and Remodeling of Glycerophosphocholine Lipids Support Cytokine Induction in Response to Fungal Patterns.',
'authors' => 'Márquez S, Fernández JJ, Mancebo C, Herrero-Sánchez C, Alonso S, Sandoval TA, Rodríguez Prados M, Cubillos-Ruiz JR, Montero O, Fernández N, Sánchez Crespo M',
'description' => '<p>Increased glycolysis parallels immune cell activation, but the role of pyruvate remains largely unexplored. We found that stimulation of dendritic cells with the fungal surrogate zymosan causes decreases of pyruvate, citrate, itaconate, and α-ketoglutarate, while increasing oxaloacetate, succinate, lactate, oxygen consumption, and pyruvate dehydrogenase activity. Expression of IL10 and IL23A (the gene encoding the p19 chain of IL-23) depended on pyruvate dehydrogenase activity. Mechanistically, pyruvate reinforced histone H3 acetylation, and acetate rescued the effect of mitochondrial pyruvate carrier inhibition, most likely because it is a substrate of the acetyl-CoA producing enzyme ACSS2. Mice lacking the receptor of the lipid mediator platelet-activating factor (PAF; 1-O-hexadecyl-2-acetyl-sn-glycero-3-phosphocholine) showed reduced production of IL-10 and IL-23 that is explained by the requirement of acetyl-CoA for PAF biosynthesis and its ensuing autocrine function. Acetyl-CoA therefore intertwines fatty acid remodeling of glycerophospholipids and energetic metabolism during cytokine induction.</p>',
'date' => '2019-04-09',
'pmid' => 'http://www.pubmed.gov/30970255',
'doi' => '10.1016/j.celrep.2019.03.033',
'modified' => '2019-08-07 09:15:46',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 33 => array(
'id' => '3693',
'name' => 'Increased Serine and One-Carbon Pathway Metabolism by PKCλ/ι Deficiency Promotes Neuroendocrine Prostate Cancer.',
'authors' => 'Reina-Campos M, Linares JF, Duran A, Cordes T, L'Hermitte A, Badur MG, Bhangoo MS, Thorson PK, Richards A, Rooslid T, Garcia-Olmo DC, Nam-Cha SY, Salinas-Sanchez AS, Eng K, Beltran H, Scott DA, Metallo CM, Moscat J, Diaz-Meco MT',
'description' => '<p>Increasingly effective therapies targeting the androgen receptor have paradoxically promoted the incidence of neuroendocrine prostate cancer (NEPC), the most lethal subtype of castration-resistant prostate cancer (PCa), for which there is no effective therapy. Here we report that protein kinase C (PKC)λ/ι is downregulated in de novo and during therapy-induced NEPC, which results in the upregulation of serine biosynthesis through an mTORC1/ATF4-driven pathway. This metabolic reprogramming supports cell proliferation and increases intracellular S-adenosyl methionine (SAM) levels to feed epigenetic changes that favor the development of NEPC characteristics. Altogether, we have uncovered a metabolic vulnerability triggered by PKCλ/ι deficiency in NEPC, which offers potentially actionable targets to prevent therapy resistance in PCa.</p>',
'date' => '2019-03-18',
'pmid' => 'http://www.pubmed.gov/30827887',
'doi' => '10.1016/j.ccell.2019.01.018',
'modified' => '2019-06-28 13:49:24',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 34 => array(
'id' => '3660',
'name' => 'Global distribution of DNA hydroxymethylation and DNA methylation in chronic lymphocytic leukemia.',
'authors' => 'Wernig-Zorc S, Yadav MP, Kopparapu PK, Bemark M, Kristjansdottir HL, Andersson PO, Kanduri C, Kanduri M',
'description' => '<p>BACKGROUND: Chronic lymphocytic leukemia (CLL) has been a good model system to understand the functional role of 5-methylcytosine (5-mC) in cancer progression. More recently, an oxidized form of 5-mC, 5-hydroxymethylcytosine (5-hmC) has gained lot of attention as a regulatory epigenetic modification with prognostic and diagnostic implications for several cancers. However, there is no global study exploring the role of 5-hydroxymethylcytosine (5-hmC) levels in CLL. Herein, using mass spectrometry and hMeDIP-sequencing, we analysed the dynamics of 5-hmC during B cell maturation and CLL pathogenesis. RESULTS: We show that naïve B-cells had higher levels of 5-hmC and 5-mC compared to non-class switched and class-switched memory B-cells. We found a significant decrease in global 5-mC levels in CLL patients (n = 15) compared to naïve and memory B cells, with no changes detected between the CLL prognostic groups. On the other hand, global 5-hmC levels of CLL patients were similar to memory B cells and reduced compared to naïve B cells. Interestingly, 5-hmC levels were increased at regulatory regions such as gene-body, CpG island shores and shelves and 5-hmC distribution over the gene-body positively correlated with degree of transcriptional activity. Importantly, CLL samples showed aberrant 5-hmC and 5-mC pattern over gene-body compared to well-defined patterns in normal B-cells. Integrated analysis of 5-hmC and RNA-sequencing from CLL datasets identified three novel oncogenic drivers that could have potential roles in CLL development and progression. CONCLUSIONS: Thus, our study suggests that the global loss of 5-hmC, accompanied by its significant increase at the gene regulatory regions, constitute a novel hallmark of CLL pathogenesis. Our combined analysis of 5-mC and 5-hmC sequencing provided insights into the potential role of 5-hmC in modulating gene expression changes during CLL pathogenesis.</p>',
'date' => '2019-01-07',
'pmid' => 'http://www.pubmed.gov/30616658',
'doi' => '10.1186/s13072‑018‑0252‑7',
'modified' => '2019-07-01 11:46:16',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 35 => array(
'id' => '3730',
'name' => 'Transcriptome-Wide Mapping 5-Methylcytosine by mC RNA Immunoprecipitation Followed by Deep Sequencing in Plant.',
'authors' => 'Gu X, Liang Z',
'description' => '<p>Transcriptome-wide mapping RNA modification is crucial to understand the distribution and function of RNA modifications. Here, we describe a protocol to transcriptome-wide mapping 5-methylcytosine (mC) in plant, by a RNA immunoprecipitation followed by deep sequencing (mC-RIP-seq) approach. The procedure includes RNA extraction, fragmentation, RNA immunoprecipitation, and library construction.</p>',
'date' => '2019-01-01',
'pmid' => 'http://www.pubmed.gov/30945199',
'doi' => '10.1007/978-1-4939-9045-0_24,',
'modified' => '2019-08-07 10:21:37',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 36 => array(
'id' => '3584',
'name' => 'Sensitivity of pituitary gonadotropes to hyperglycemia leads to epigenetic aberrations and reduced follicle-stimulating hormone levels.',
'authors' => 'Feldman A, Saleh A, Pnueli L, Qiao S, Shlomi T, Boehm U, Melamed P',
'description' => '<p>The connection between metabolism and reproductive function is well recognized, and we hypothesized that the pituitary gonadotropes, which produce luteinizing hormone and follicle-stimulating hormone (FSH), mediate some of the effects directly via insulin-independent glucose transporters, which allow continued glucose metabolism during hyperglycemia. We found that glucose transporter 1 is the predominant glucose transporter in primary gonadotropes and a gonadotrope precursor-derived cell line, and both are responsive to culture in high glucose; moreover, metabolite levels were altered in the cell line. Several of the affected metabolites are cofactors for chromatin-modifying enzymes, and in the gonadotrope precursor-derived cell line, we recorded global changes in histone acetylation and methylation, decreased DNA methylation, and increased hydroxymethylation, some of which did not revert to basal levels after cells were returned to normal glucose. Despite this weakening of epigenetic-mediated repression seen in the model cell line, FSH β-subunit ( Fshb) mRNA levels in primary gonadotropes were significantly reduced, apparently due in part to increased autocrine/paracrine effects of inhibin. However, unlike thioredoxin interacting protein and inhibin subunit α, Fshb mRNA levels did not recover after the return of cells to normal glucose. The effect on Fshb expression was also seen in 2 hyperglycemic mouse models, and levels of circulating FSH, required for follicle growth and development, were reduced. Thus, hyperglycemia seems to target the pituitary gonadotropes directly, and the likely extensive epigenetic changes are sensed acutely by Fshb. This scenario would explain clinical findings in which, even after restoration of optimal blood glucose levels, fertility often remains adversely affected. However, the relative accessibility of the pituitary provides a possible target for treatment, particularly crucial in the young in which hyperglycemia is increasingly common and fertility most relevant.-Feldman, A., Saleh, A., Pnueli, L., Qiao, S., Shlomi, T., Boehm, U., Melamed, P. Sensitivity of pituitary gonadotropes to hyperglycemia leads to epigenetic aberrations and reduced follicle-stimulating hormone levels.</p>',
'date' => '2018-12-27',
'pmid' => 'http://www.pubmed.gov/30074825',
'doi' => '10.1096/fj.201800943R',
'modified' => '2019-04-17 15:48:51',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 37 => array(
'id' => '3430',
'name' => 'Sensitive tumour detection and classification using plasma cell-free DNA methylomes.',
'authors' => 'Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, Kis O, Zhao Z, Spreafico A, Medina TDS, Wang Y, Roulois D, Ettayebi I, Chen Z, Chow S, Murphy T, Arruda A, O'Kane GM, Liu J, Mansour M, McPher',
'description' => '<p>The use of liquid biopsies for cancer detection and management is rapidly gaining prominence. Current methods for the detection of circulating tumour DNA involve sequencing somatic mutations using cell-free DNA, but the sensitivity of these methods may be low among patients with early-stage cancer given the limited number of recurrent mutations. By contrast, large-scale epigenetic alterations-which are tissue- and cancer-type specific-are not similarly constrained and therefore potentially have greater ability to detect and classify cancers in patients with early-stage disease. Here we develop a sensitive, immunoprecipitation-based protocol to analyse the methylome of small quantities of circulating cell-free DNA, and demonstrate the ability to detect large-scale DNA methylation changes that are enriched for tumour-specific patterns. We also demonstrate robust performance in cancer detection and classification across an extensive collection of plasma samples from several tumour types. This work sets the stage to establish biomarkers for the minimally invasive detection, interception and classification of early-stage cancers based on plasma cell-free DNA methylation patterns.</p>',
'date' => '2018-11-14',
'pmid' => 'http://www.pubmed.gov/30429608',
'doi' => '10.1038/s41586-018-0703-0',
'modified' => '2019-06-11 16:22:54',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 38 => array(
'id' => '3421',
'name' => 'Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate.',
'authors' => 'Sardina JL, Collombet S, Tian TV, Gómez A, Di Stefano B, Berenguer C, Brumbaugh J, Stadhouders R, Segura-Morales C, Gut M, Gut IG, Heath S, Aranda S, Di Croce L, Hochedlinger K, Thieffry D, Graf T',
'description' => '<p>Here, we report DNA methylation and hydroxymethylation dynamics at nucleotide resolution using C/EBPα-enhanced reprogramming of B cells into induced pluripotent cells (iPSCs). We observed successive waves of hydroxymethylation at enhancers, concomitant with a decrease in DNA methylation, suggesting active demethylation. Consistent with this finding, ablation of the DNA demethylase Tet2 almost completely abolishes reprogramming. C/EBPα, Klf4, and Tfcp2l1 each interact with Tet2 and recruit the enzyme to specific DNA sites. During reprogramming, some of these sites maintain high levels of 5hmC, and enhancers and promoters of key pluripotency factors become demethylated as early as 1 day after Yamanaka factor induction. Surprisingly, methylation changes precede chromatin opening in distinct chromatin regions, including Klf4 bound sites, revealing a pioneer factor activity associated with alternation in DNA methylation. Rapid changes in hydroxymethylation similar to those in B cells were also observed during compound-accelerated reprogramming of fibroblasts into iPSCs, highlighting the generality of our observations.</p>',
'date' => '2018-11-01',
'pmid' => 'http://www.pubmed.gov/30220521',
'doi' => '10.1016/j.stem.2018.08.016',
'modified' => '2018-12-31 11:16:24',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 39 => array(
'id' => '3409',
'name' => 'Oxidative stress in sperm affects the epigenetic reprogramming in early embryonic development.',
'authors' => 'Wyck S, Herrera C, Requena CE, Bittner L, Hajkova P, Bollwein H, Santoro R',
'description' => '<p>BACKGROUND: Reactive oxygen species (ROS)-induced oxidative stress is well known to play a major role in male infertility. Sperm are sensitive to ROS damaging effects because as male germ cells form mature sperm they progressively lose the ability to repair DNA damage. However, how oxidative DNA lesions in sperm affect early embryonic development remains elusive. RESULTS: Using cattle as model, we show that fertilization using sperm exposed to oxidative stress caused a major developmental arrest at the time of embryonic genome activation. The levels of DNA damage response did not directly correlate with the degree of developmental defects. The early cellular response for DNA damage, γH2AX, is already present at high levels in zygotes that progress normally in development and did not significantly increase at the paternal genome containing oxidative DNA lesions. Moreover, XRCC1, a factor implicated in the last step of base excision repair (BER) pathway, was recruited to the damaged paternal genome, indicating that the maternal BER machinery can repair these DNA lesions induced in sperm. Remarkably, the paternal genome with oxidative DNA lesions showed an impairment of zygotic active DNA demethylation, a process that previous studies linked to BER. Quantitative immunofluorescence analysis and ultrasensitive LC-MS-based measurements revealed that oxidative DNA lesions in sperm impair active DNA demethylation at paternal pronuclei, without affecting 5-hydroxymethylcytosine (5hmC), a 5-methylcytosine modification that has been implicated in paternal active DNA demethylation in mouse zygotes. Thus, other 5hmC-independent processes are implicated in active DNA demethylation in bovine embryos. The recruitment of XRCC1 to damaged paternal pronuclei indicates that oxidative DNA lesions drive BER to repair DNA at the expense of DNA demethylation. Finally, this study highlighted striking differences in DNA methylation dynamics between bovine and mouse zygotes that will facilitate the understanding of the dynamics of DNA methylation in early development. CONCLUSIONS: The data demonstrate that oxidative stress in sperm has an impact not only on DNA integrity but also on the dynamics of epigenetic reprogramming, which may harm the paternal genetic and epigenetic contribution to the developing embryo and affect embryo development and embryo quality.</p>',
'date' => '2018-10-17',
'pmid' => 'http://www.pubmed.gov/30333056',
'doi' => '10.1186/s13072-018-0224-y',
'modified' => '2018-11-09 11:10:58',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 40 => array(
'id' => '3404',
'name' => 'Integrated analysis of DNA methylation profiling and gene expression profiling identifies novel markers in lung cancer in Xuanwei, China.',
'authors' => 'Wang J, Duan Y, Meng QH, Gong R, Guo C, Zhao Y, Zhang Y',
'description' => '<p>BACKGROUND: Aberrant DNA methylation occurs frequently in cancer. The aim of this study was to identify novel methylation markers in lung cancer in Xuanwei, China, through integrated genome-wide DNA methylation and gene expression studies. METHODS: Differentially methylated regions (DMRs) and differentially expressed genes (DEGs) were detected on 10 paired lung cancer tissues and noncancerous lung tissues by methylated DNA immunoprecipitation combined with microarray (MeDIP-chip) and gene expression microarray analyses, respectively. Integrated analysis of DMRs and DEGs was performed to screen out candidate methylation-related genes. Both methylation and expression changes of the candidate genes were further validated and analyzed. RESULTS: Compared with normal lung tissues, lung cancer tissues expressed a total of 6,899 DMRs, including 5,788 hypermethylated regions and 1,111 hypomethylated regions. Integrated analysis of DMRs and DEGs identified 45 tumor-specific candidate genes: 38 genes whose DMRs were hypermethylated and expression was downregulated, and 7 genes whose DMRs were hypomethylated and expression was upregulated. The methylation and expression validation results identified 4 candidate genes (STXBP6, BCL6B, FZD10, and HSPB6) that were significantly hypermethylated and downregulated in most of the tumor tissues compared with the noncancerous lung tissues. CONCLUSIONS: This integrated analysis of genome-wide DNA methylation and gene expression in lung cancer in Xuanwei revealed several genes regulated by promoter methylation that have not been described in lung cancer before. These results provide new insight into the carcinogenesis of lung cancer in Xuanwei and represent promising new diagnostic and therapeutic targets.</p>',
'date' => '2018-10-04',
'pmid' => 'http://www.pubmed.gov/30286088',
'doi' => '10.1371/journal.pone.0203155',
'modified' => '2018-11-09 11:24:21',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 41 => array(
'id' => '3417',
'name' => 'mGlu1 Receptors Monopolize the Synaptic Control of Cerebellar Purkinje Cells by Epigenetically Down-Regulating mGlu5 Receptors.',
'authors' => 'Notartomaso S, Nakao H, Mascio G, Scarselli P, Cannella M, Zappulla C, Madonna M, Motolese M, Gradini R, Liberatore F, Zonta M, Carmignoto G, Battaglia G, Bruno V, Watanabe M, Aiba A, Nicoletti F',
'description' => '<p>In cerebellar Purkinje cells (PCs) type-1 metabotropic glutamate (mGlu1) receptors play a key role in motor learning and drive the refinement of synaptic innervation during postnatal development. The cognate mGlu5 receptor is absent in mature PCs and shows low expression levels in the adult cerebellar cortex. Here we found that mGlu5 receptors were heavily expressed by PCs in the early postnatal life, when mGlu1α receptors were barely detectable. The developmental decline of mGlu5 receptors coincided with the appearance of mGlu1α receptors in PCs, and both processes were associated with specular changes in CpG methylation in the corresponding gene promoters. It was the mGlu1 receptor that drove the elimination of mGlu5 receptors from PCs, as shown by data obtained with conditional mGlu1α receptor knockout mice and with targeted pharmacological treatments during critical developmental time windows. The suppressing activity of mGlu1 receptors on mGlu5 receptor was maintained in mature PCs, suggesting that expression of mGlu1α and mGlu5 receptors is mutually exclusive in PCs. These findings add complexity to the the finely tuned mechanisms that regulate PC biology during development and in the adult life and lay the groundwork for an in-depth analysis of the role played by mGlu5 receptors in PC maturation.</p>',
'date' => '2018-09-06',
'pmid' => 'http://www.pubmed.gov/30190524',
'doi' => '10.1038/s41598-018-31369-7',
'modified' => '2018-12-31 11:36:04',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 42 => array(
'id' => '3640',
'name' => 'Determination of the presence of 5-methylcytosine in Paramecium tetraurelia.',
'authors' => 'Singh A, Vancura A, Woycicki RK, Hogan DJ, Hendrick AG, Nowacki M',
'description' => '<p>5-methylcytosine DNA methylation regulates gene expression and developmental programming in a broad range of eukaryotes. However, its presence and potential roles in ciliates, complex single-celled eukaryotes with germline-somatic genome specialization via nuclear dimorphism, are largely uncharted. While canonical cytosine methyltransferases have not been discovered in published ciliate genomes, recent studies performed in the stichotrichous ciliate Oxytricha trifallax suggest de novo cytosine methylation during macronuclear development. In this study, we applied bisulfite genome sequencing, DNA mass spectrometry and antibody-based fluorescence detection to investigate the presence of DNA methylation in Paramecium tetraurelia. While the antibody-based methods suggest cytosine methylation, DNA mass spectrometry and bisulfite sequencing reveal that levels are actually below the limit of detection. Our results suggest that Paramecium does not utilize 5-methylcytosine DNA methylation as an integral part of its epigenetic arsenal.</p>',
'date' => '2018-06-07',
'pmid' => 'http://www.pubmed.gov/30379964',
'doi' => '10.1371/journal.',
'modified' => '2019-06-07 10:22:28',
'created' => '2019-06-06 12:11:18',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 43 => array(
'id' => '3458',
'name' => 'Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.',
'authors' => 'Li T, Wang L, Du Y, Xie S, Yang X, Lian F, Zhou Z, Qian C',
'description' => '<p>UHRF1 plays multiple roles in regulating DNMT1-mediated DNA methylation maintenance during DNA replication. The UHRF1 C-terminal RING finger functions as an ubiquitin E3 ligase to establish histone H3 ubiquitination at Lys18 and/or Lys23, which is subsequently recognized by DNMT1 to promote its localization onto replication foci. Here, we present the crystal structure of DNMT1 RFTS domain in complex with ubiquitin and highlight a unique ubiquitin binding mode for the RFTS domain. We provide evidence that UHRF1 N-terminal ubiquitin-like domain (UBL) also binds directly to DNMT1. Despite sharing a high degree of structural similarity, UHRF1 UBL and ubiquitin bind to DNMT1 in a very distinct fashion and exert different impacts on DNMT1 enzymatic activity. We further show that the UHRF1 UBL-mediated interaction between UHRF1 and DNMT1, and the binding of DNMT1 to ubiquitinated histone H3 that is catalyzed by UHRF1 RING domain are critical for the proper subnuclear localization of DNMT1 and maintenance of DNA methylation. Collectively, our study adds another layer of complexity to the regulatory mechanism of DNMT1 activation by UHRF1 and supports that individual domains of UHRF1 participate and act in concert to maintain DNA methylation patterns.</p>',
'date' => '2018-04-06',
'pmid' => 'http://www.pubmed.gov/29471350',
'doi' => '10.1093/nar/gky104',
'modified' => '2019-02-15 21:14:42',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 44 => array(
'id' => '3475',
'name' => 'Epigenetics and early domestication: differences in hypothalamic DNA methylation between red junglefowl divergently selected for high or low fear of humans.',
'authors' => 'Bélteky J, Agnvall B, Bektic L, Höglund A, Jensen P, Guerrero-Bosagna C',
'description' => '<p>BACKGROUND: Domestication of animals leads to large phenotypic alterations within a short evolutionary time-period. Such alterations are caused by genomic variations, yet the prevalence of modified traits is higher than expected if they were caused only by classical genetics and mutations. Epigenetic mechanisms may also be important in driving domesticated phenotypes such as behavior traits. Gene expression can be modulated epigenetically by mechanisms such as DNA methylation, resulting in modifications that are not only variable and susceptible to environmental stimuli, but also sometimes transgenerationally stable. To study such mechanisms in early domestication, we used as model two selected lines of red junglefowl (ancestors of modern chickens) that were bred for either high or low fear of humans over five generations, and investigated differences in hypothalamic DNA methylation between the two populations. RESULTS: Twenty-two 1-kb windows were differentially methylated between the two selected lines at p < 0.05 after false discovery rate correction. The annotated functions of the genes within these windows indicated epigenetic regulation of metabolic and signaling pathways, which agrees with the changes in gene expression that were previously reported for the same tissue and animals. CONCLUSIONS: Our results show that selection for an important domestication-related behavioral trait such as tameness can cause divergent epigenetic patterns within only five generations, and that these changes could have an important role in chicken domestication.</p>',
'date' => '2018-04-02',
'pmid' => 'http://www.pubmed.gov/29609558',
'doi' => '10.1186/s12711-018-0384-z',
'modified' => '2019-02-15 20:32:37',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 45 => array(
'id' => '3393',
'name' => 'Copper induces expression and methylation changes of early development genes in Crassostrea gigas embryos.',
'authors' => 'Sussarellu R, Lebreton M, Rouxel J, Akcha F, Rivière G',
'description' => '<p>Copper contamination is widespread along coastal areas and exerts adverse effects on marine organisms such as mollusks. In the Pacific oyster, copper induces severe developmental abnormalities during early life stages; however, the underlying molecular mechanisms are largely unknown. This study aims to better understand whether the embryotoxic effects of copper in Crassostrea gigas could be mediated by alterations in gene expression, and the putative role of DNA methylation, which is known to contribute to gene regulation in early embryo development. For that purpose, oyster embryos were exposed to 4 nominal copper concentrations (0.1, 1, 10 and 20 μg L Cu) during early development assays. Embryotoxicity was monitored through the oyster embryo-larval bioassay at the D-larva stage 24 h post fertilization (hpf) and genotoxicity at gastrulation 7 hpf. In parallel, the relative expression of 15 genes encoding putative homeotic, biomineralization and DNA methylation proteins was measured at three developmental stages (3 hpf morula stage, 7 hpf gastrula stage, 24 hpf D-larvae stage) using RT-qPCR. Global DNA content in methylcytosine and hydroxymethylcytosine were measured by HPLC and gene-specific DNA methylation levels were monitored using MeDIP-qPCR. A significant increase in larval abnormalities was observed from copper concentrations of 10 μg L, while significant genotoxic effects were detected at 1 μg L and above. All the selected genes presented a stage-dependent expression pattern, which was impaired for some homeobox and DNA methylation genes (Notochord, HOXA1, HOX2, Lox5, DNMT3b and CXXC-1) after copper exposure. While global DNA methylation (5-methylcytosine) at gastrula stage didn't show significant changes between experimental conditions, 5-hydroxymethylcytosine, its degradation product, decreased upon copper treatment. The DNA methylation of exons and the transcript levels were correlated in control samples for HOXA1 but such a correlation was diminished following copper exposure. The methylation level of some specific gene regions (HoxA1, Hox2, Engrailed2 and Notochord) displayed changes upon copper exposure. Such changes were gene and exon-specific and no obvious global trends could be identified. Our study suggests that the embryotoxic effects of copper in oysters could involve homeotic gene expression impairment possibly by changing DNA methylation levels.</p>',
'date' => '2018-03-01',
'pmid' => 'http://www.pubmed.gov/29353135',
'doi' => '10.1016/j.aquatox.2018.01.001',
'modified' => '2018-11-09 12:21:38',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 46 => array(
'id' => '3448',
'name' => 'Aberrant methylated key genes of methyl group metabolism within the molecular etiology of urothelial carcinogenesis.',
'authors' => 'Erichsen L, Ghanjati F, Beermann A, Poyet C, Hermanns T, Schulz WA, Seifert HH, Wild PJ, Buser L, Kröning A, Braunstein S, Anlauf M, Jankowiak S, Hassan M, Bendhack ML, Araúzo-Bravo MJ, Santourlidis S',
'description' => '<p>Urothelial carcinoma (UC), the most common cancer of the urinary bladder causes severe morbidity and mortality, e.g. about 40.000 deaths in the EU annually, and incurs considerable costs for the health system due to the need for prolonged treatments and long-term monitoring. Extensive aberrant DNA methylation is described to prevail in urothelial carcinoma and is thought to contribute to genetic instability, altered gene expression and tumor progression. However, it is unknown how this epigenetic alteration arises during carcinogenesis. Intact methyl group metabolism is required to ensure maintenance of cell-type specific methylomes and thereby genetic integrity and proper cellular function. Here, using two independent techniques for detecting DNA methylation, we observed DNA hypermethylation of the 5'-regulatory regions of the key methyl group metabolism genes ODC1, AHCY and MTHFR in early urothelial carcinoma. These hypermethylation events are associated with genome-wide DNA hypomethylation which is commonly associated with genetic instability. We therefore infer that hypermethylation of methyl group metabolism genes acts in a feed-forward cycle to promote additional DNA methylation changes and suggest a new hypothesis on the molecular etiology of urothelial carcinoma.</p>',
'date' => '2018-02-22',
'pmid' => 'http://www.pubmed.gov/29472622',
'doi' => '10.1038/s41598-018-21932-7',
'modified' => '2019-02-15 21:31:04',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 47 => array(
'id' => '3383',
'name' => 'Genome-wide analysis of day/night DNA methylation differences in Populus nigra.',
'authors' => 'Ding C.J. et al.',
'description' => '<p>DNA methylation is an important mechanism of epigenetic modification. Methylation changes during stress responses and developmental processes have been well studied; however, their role in plant adaptation to the day/night cycle is poorly understood. In this study, we detected global methylation patterns in leaves of the black poplar Populus nigra 'N46' at 8:00 and 24:00 by methylated DNA immunoprecipitation sequencing (MeDIP-seq). We found 10,027 and 10,242 genes to be methylated in the 8:00 and 24:00 samples, respectively. The methylated genes appeared to be involved in multiple biological processes, molecular functions, and cellular components, suggesting important roles for DNA methylation in poplar cells. Comparing the 8:00 and 24:00 samples, only 440 differentially methylated regions (DMRs) overlapped with genic regions, including 193 hyper- and 247 hypo-methylated DMRs, and may influence the expression of 137 downstream genes. Most hyper-methylated genes were associated with transferase activity, kinase activity, and phosphotransferase activity, whereas most hypo-methylated genes were associated with protein binding, ATP binding, and adenyl ribonucleotide binding, suggesting that different biological processes were activated during the day and night. Our results indicated that methylated genes were prevalent in the poplar genome, but that only a few of these participated in diurnal gene expression regulation.</p>',
'date' => '2018-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29293569',
'doi' => '',
'modified' => '2018-08-07 09:45:38',
'created' => '2018-08-07 09:45:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 48 => array(
'id' => '3384',
'name' => 'Obligatory and facilitative allelic variation in the DNA methylome within common disease-associated loci',
'authors' => 'Bell C.G. et al.',
'description' => '<p>Integrating epigenetic data with genome-wide association study (GWAS) results can reveal disease mechanisms. The genome sequence itself also shapes the epigenome, with CpG density and transcription factor binding sites (TFBSs) strongly encoding the DNA methylome. Therefore, genetic polymorphism impacts on the observed epigenome. Furthermore, large genetic variants alter epigenetic signal dosage. Here, we identify DNA methylation variability between GWAS-SNP risk and non-risk haplotypes. In three subsets comprising 3128 MeDIP-seq peripheral-blood DNA methylomes, we find 7173 consistent and functionally enriched Differentially Methylated Regions. 36.8% can be attributed to common non-SNP genetic variants. CpG-SNPs, as well as facilitative TFBS-motifs, are also enriched. Highlighting their functional potential, CpG-SNPs strongly associate with allele-specific DNase-I hypersensitivity sites. Our results demonstrate strong DNA methylation allelic differences driven by obligatory or facilitative genetic effects, with potential direct or regional disease-related repercussions. These allelic variations require disentangling from pure tissue-specific modifications, may influence array studies, and imply underestimated population variability in current reference epigenomes.</p>',
'date' => '2018-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29295990',
'doi' => '',
'modified' => '2018-08-07 10:13:12',
'created' => '2018-08-07 10:13:12',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 49 => array(
'id' => '3508',
'name' => 'Analysis of DNA methylome and transcriptome profiling following Gibberellin A3 (GA3) foliar application in Nicotiana tabacum L.',
'authors' => 'Manoharlal Raman, Saiprasad G. V. S., Kaikala Vinay, Suresh Kumar R., Kovařík Ales',
'description' => '<p>The present work investigated a comprehensive genome-wide landscape of DNA methylome and its relationship with transcriptome upon gibberellin A3 (GA3) foliar application under practical field conditions in solanaceae model, Nicotiana tabacum L. Methylated DNA Immunoprecipitation-Sequencing (MeDIP-Seq) analysis uncovered over 82% (18,456) of differential methylated regions (DMRs) in intergenic-region. Within protein-coding region, 2339 and 1685 of identified DMRs were observed in genebody- and promoter-region, respectively. Microarray study revealed 7032 differential expressed genes (DEGs) with 3507 and 3525 genes displaying upand down-regulation, respectively. Integration analysis revealed 520 unique non-redundant annotated DMRs overlapping with DEGs. Our results indicated that GA3 induced DNA hypo- as well as hyper-methylation were associated with both gene-silencing and -activation. No complete biasness or correlation was observed in either of the promoter- or genebody-regions, which otherwise showed an overall trend towards GA3 induced global DNA hypo-methylation. Taken together, our results suggested that differential DNA methylation mediated by GA3 may only play a permissive role in regulating the gene expression.</p>',
'date' => '2018-01-01',
'pmid' => 'https://link.springer.com/article/10.1007/s40502-018-0393-5',
'doi' => '10.1007/s40502-018-0393-5',
'modified' => '2022-05-18 18:43:47',
'created' => '2019-02-27 12:54:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 50 => array(
'id' => '3297',
'name' => '5-Methylcytosine RNA Methylation in Arabidopsis Thaliana',
'authors' => 'Cui X. et al.',
'description' => '<p>5-Methylcytosine (m<sup>5</sup>C) is a well-characterized DNA modification, and is also predominantly reported in abundant non-coding RNAs in both prokaryotes and eukaryotes. However, the distribution and biological functions of m<sup>5</sup>C in plant mRNAs remain largely unknown. Here, we report transcriptome-wide profiling of RNA m<sup>5</sup>C in Arabidopsis thaliana by applying m<sup>5</sup>C RNA immunoprecipitation followed by a deep-sequencing approach (m<sup>5</sup>C-RIP-seq). LC-MS/MS and dot blot analyses reveal a dynamic pattern of m<sup>5</sup>C mRNA modification in various tissues and at different developmental stages. m<sup>5</sup>C-RIP-seq analysis identified 6045 m<sup>5</sup>C peaks in 4465 expressed genes in young seedlings. We found that m<sup>5</sup>C is enriched in coding sequences with two peaks located immediately after start codons and before stop codons, and is associated with mRNAs with low translation activity. We further demonstrated that an RNA (cytosine-5)-methyltransferase, tRNA-specific methyltransferase 4B (TRM4B), exhibits m<sup>5</sup>C RNA methyltransferase activity. Mutations in TRM4B display defects in root development and decreased m<sup>5</sup>C peaks. TRM4B affects the transcript levels of the genes involved in root development, which is positively correlated with their mRNA stability and m<sup>5</sup>C levels. Our results suggest that m<sup>5</sup>C in mRNA is a new epitranscriptome marker inArabidopsis, and that regulation of this modification is an integral part of gene regulatory networks underlying plant development.</p>',
'date' => '2017-11-06',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28965832',
'doi' => '',
'modified' => '2017-12-04 11:10:34',
'created' => '2017-12-04 11:10:34',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 51 => array(
'id' => '3220',
'name' => 'Maternal obesity programs increased leptin gene expression in rat male offspring via epigenetic modifications in a depot-specific manner',
'authors' => 'Lecoutre S. et al.',
'description' => '<div class="">
<h4>OBJECTIVE:</h4>
<p><abstracttext label="OBJECTIVE" nlmcategory="OBJECTIVE">According to the Developmental Origin of Health and Disease (DOHaD) concept, maternal obesity and accelerated growth in neonates predispose offspring to white adipose tissue (WAT) accumulation. In rodents, adipogenesis mainly develops during lactation. The mechanisms underlying the phenomenon known as developmental programming remain elusive. We previously reported that adult rat offspring from high-fat diet-fed dams (called HF) exhibited hypertrophic adipocyte, hyperleptinemia and increased leptin mRNA levels in a depot-specific manner. We hypothesized that leptin upregulation occurs via epigenetic malprogramming, which takes place early during development of WAT.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">As a first step, we identified <i>in silico</i> two potential enhancers located upstream and downstream of the leptin transcription start site that exhibit strong dynamic epigenomic remodeling during adipocyte differentiation. We then focused on epigenetic modifications (methylation, hydroxymethylation, and histone modifications) of the promoter and the two potential enhancers regulating leptin gene expression in perirenal (pWAT) and inguinal (iWAT) fat pads of HF offspring during lactation (postnatal days 12 (PND12) and 21 (PND21)) and in adulthood.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">PND12 is an active period for epigenomic remodeling in both deposits especially in the upstream enhancer, consistent with leptin gene induction during adipogenesis. Unlike iWAT, some of these epigenetic marks were still observable in pWAT of weaned HF offspring. Retained marks were only visible in pWAT of 9-month-old HF rats that showed a persistent "expandable" phenotype.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Consistent with the DOHaD hypothesis, persistent epigenetic remodeling occurs at regulatory regions especially within intergenic sequences, linked to higher leptin gene expression in adult HF offspring in a depot-specific manner.</abstracttext></p>
</div>',
'date' => '2017-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5518658/',
'doi' => '',
'modified' => '2017-08-18 13:56:40',
'created' => '2017-08-18 13:56:40',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 52 => array(
'id' => '3204',
'name' => 'Increased 5-hydroxymethylation levels in the hippocampus of rat extinguished from cocaine self-administration',
'authors' => 'Sadakierska-Chudy A. et al.',
'description' => '<p>Drug craving and relapse risk during abstinence from cocaine are thought to be caused by persistent changes in transcription and chromatin regulation. Although several brain regions are involved in these processes, the hippocampus seems to play an important role in context-evoked craving and drug-seeking behavior. Only a few studies have examined epigenetic alterations during a period of cocaine abstinence. To investigate the effects of cocaine abstinence on DNA methylation and gene expression, rats that self-administered the drug underwent cocaine abstinence in two time points with extinction training. During the cocaine extinction, we observed elevated global 5-hydroxymethylcytosine(5-hmC) levels with a concurrent increase in Tet3 transcript levels. Moreover, we did not find significant alterations in the levels of Tet3 mRNA and 5-hmC in rats subjected to cocaine abstinence without extinction training. Additionally, our findings demonstrated that the expression of Tet3 target genes was activated. Besides, altered DNA methylation was detected at promoter regions of miRNAs, such as miR-30d and miR-let7i. Further in silico analysis provided evidence that these two molecules targeted the 3' UTR region of the Tet3 gene and thus may contribute to its post-transcriptional regulation. This study has presented novel findings in the hippocampus of rats that underwent extinction training following cocaine self-administration. The alterations in the Tet3 gene expression and the level of 5-hmC may play an important role in extinction learning and the reduction of subsequent cocaine seeking.</p>',
'date' => '2017-07-27',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28422379',
'doi' => '',
'modified' => '2017-07-03 10:21:48',
'created' => '2017-07-03 10:21:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 53 => array(
'id' => '3225',
'name' => 'The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation',
'authors' => 'Leone S. et al.',
'description' => '<p>Long non-coding RNAs (lncRNAs) have been implicated in the regulation of chromatin conformation and epigenetic patterns. lncRNA expression levels are widely taken as an indicator for functional properties. However, the role of RNA processing in modulating distinct features of the same lncRNA is less understood. The establishment of heterochromatin at rRNA genes depends on the processing of IGS-rRNA into pRNA, a reaction that is impaired in embryonic stem cells (ESCs) and activated only upon differentiation. The production of mature pRNA is essential since it guides the repressor TIP5 to rRNA genes, and IGS-rRNA abolishes this process. Through screening for IGS-rRNA-binding proteins, we here identify the RNA helicase DHX9 as a regulator of pRNA processing. DHX9 binds to rRNA genes only upon ESC differentiation and its activity guides TIP5 to rRNA genes and establishes heterochromatin. Remarkably, ESCs depleted of DHX9 are unable to differentiate and this phenotype is reverted by the addition of pRNA, whereas providing IGS-rRNA and pRNA mutants deficient for TIP5 binding are not sufficient. Our results reveal insights into lncRNA biogenesis during development and support a model in which the state of rRNA gene chromatin is part of the regulatory network that controls exit from pluripotency and initiation of differentiation pathways.</p>',
'date' => '2017-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28588071',
'doi' => '',
'modified' => '2017-08-22 13:52:28',
'created' => '2017-08-22 13:52:28',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 54 => array(
'id' => '3233',
'name' => 'Pramel7 mediates ground-state pluripotency through proteasomal-epigenetic combined pathways.',
'authors' => 'Graf U. et al.',
'description' => '<p>Naive pluripotency is established in preimplantation epiblast. Embryonic stem cells (ESCs) represent the immortalization of naive pluripotency. 2i culture has optimized this state, leading to a gene signature and DNA hypomethylation closely comparable to preimplantation epiblast, the developmental ground state. Here we show that Pramel7 (PRAME-like 7), a protein highly expressed in the inner cell mass (ICM) but expressed at low levels in ESCs, targets for proteasomal degradation UHRF1, a key factor for DNA methylation maintenance. Increasing Pramel7 expression in serum-cultured ESCs promotes a preimplantation epiblast-like gene signature, reduces UHRF1 levels and causes global DNA hypomethylation. Pramel7 is required for blastocyst formation and its forced expression locks ESCs in pluripotency. Pramel7/UHRF1 expression is mutually exclusive in ICMs whereas Pramel7-knockout embryos express high levels of UHRF1. Our data reveal an as-yet-unappreciated dynamic nature of DNA methylation through proteasome pathways and offer insights that might help to improve ESC culture to reproduce in vitro the in vivo ground-state pluripotency.</p>',
'date' => '2017-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28604677',
'doi' => '',
'modified' => '2017-08-24 09:57:28',
'created' => '2017-08-24 09:57:28',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 55 => array(
'id' => '3200',
'name' => 'CHD4 Has Oncogenic Functions in Initiating and Maintaining Epigenetic Suppression of Multiple Tumor Suppressor Genes',
'authors' => 'Xia L. et al.',
'description' => '<p>An oncogenic role for CHD4, a NuRD component, is defined for initiating and supporting tumor suppressor gene (TSG) silencing in human colorectal cancer. CHD4 recruits repressive chromatin proteins to sites of DNA damage repair, including DNA methyltransferases where it imposes de novo DNA methylation. At TSGs, CHD4 retention helps maintain DNA hypermethylation-associated transcriptional silencing. CHD4 is recruited by the excision repair protein OGG1 for oxidative damage to interact with the damage-induced base 8-hydroxydeoxyguanosine (8-OHdG), while ZMYND8 recruits it to double-strand breaks. CHD4 knockdown activates silenced TSGs, revealing their role for blunting colorectal cancer cell proliferation, invasion, and metastases. High CHD4 and 8-OHdG levels plus low expression of TSGs strongly correlates with early disease recurrence and decreased overall survival.</p>',
'date' => '2017-05-08',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28486105',
'doi' => '',
'modified' => '2017-07-03 09:56:32',
'created' => '2017-07-03 09:56:32',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 56 => array(
'id' => '3170',
'name' => 'Critical threshold levels of DNA methyltransferase 1 are required to maintain DNA methylation across the genome in human cancer cells.',
'authors' => 'Cai Y. et al.',
'description' => '<p>Reversing DNA methylation abnormalities and associated gene silencing, through inhibiting DNA methyltransferases (DNMTs) is an important potential cancer therapy paradigm. Maximizing this potential requires defining precisely how these enzymes maintain genome-wide, cancer-specific DNA methylation. To date, there is incomplete understanding of precisely how the three DNMTs, 1, 3A, and 3B, interact for maintaining DNA methylation abnormalities in cancer. By combining genetic and shRNA depletion strategies, we define not only a dominant role for DNA methyltransferase 1 (DNMT1) but also distinct roles of 3A and 3B in genome-wide DNA methylation maintenance. Lowering DNMT1 below a threshold level is required for maximal loss of DNA methylation at all genomic regions, including gene body and enhancer regions, and for maximally reversing abnormal promoter DNA hypermethylation and associated gene silencing to reexpress key genes. It is difficult to reach this threshold with patient-tolerable doses of current DNMT inhibitors (DNMTIs). We show that new approaches, like decreasing the DNMT targeting protein, UHRF1, can augment the DNA demethylation capacities of existing DNA methylation inhibitors for fully realizing their therapeutic potential.</p>',
'date' => '2017-04-27',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28232479',
'doi' => '',
'modified' => '2017-05-10 15:31:12',
'created' => '2017-05-10 15:31:12',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 57 => array(
'id' => '3228',
'name' => 'Regulation of DNA demethylation by the XPC DNA repair complex in somatic and pluripotent stem cells.',
'authors' => 'Ho J.J. et al.',
'description' => '<p>Faithful resetting of the epigenetic memory of a somatic cell to a pluripotent state during cellular reprogramming requires DNA methylation to silence somatic gene expression and dynamic DNA demethylation to activate pluripotency gene transcription. The removal of methylated cytosines requires the base excision repair enzyme TDG, but the mechanism by which TDG-dependent DNA demethylation occurs in a rapid and site-specific manner remains unclear. Here we show that the XPC DNA repair complex is a potent accelerator of global and locus-specific DNA demethylation in somatic and pluripotent stem cells. XPC cooperates with TDG genome-wide to stimulate the turnover of essential intermediates by overcoming slow TDG-abasic product dissociation during active DNA demethylation. We further establish that DNA demethylation induced by XPC expression in somatic cells overcomes an early epigenetic barrier in cellular reprogramming and facilitates the generation of more robust induced pluripotent stem cells, characterized by enhanced pluripotency-associated gene expression and self-renewal capacity. Taken together with our previous studies establishing the XPC complex as a transcriptional coactivator, our findings underscore two distinct but complementary mechanisms by which XPC influences gene regulation by coordinating efficient TDG-mediated DNA demethylation along with active transcription during somatic cell reprogramming.</p>',
'date' => '2017-04-05',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28512237',
'doi' => '',
'modified' => '2017-08-23 14:20:13',
'created' => '2017-08-23 14:20:13',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 58 => array(
'id' => '3142',
'name' => 'Epigenetic regulation of RELN and GAD1 in the frontal cortex (FC) of autism spectrum disorder (ASD) subjects',
'authors' => 'Zhubi A. et al.',
'description' => '<p>Both Reelin (RELN) and glutamate decarboxylase 67 (GAD1) have been implicated in the pathophysiology of Autism Spectrum Disorders (ASD). We have previously shown that both mRNAs are reduced in the cerebella (CB) of ASD subjects through a mechanism that involves increases in the amounts of MECP2 binding to the corresponding promoters. In the current study, we examined the expression of RELN, GAD1, GAD2, and several other mRNAs implicated in this disorder in the frontal cortices (FC) of ASD and CON subjects. We also focused on the role that epigenetic processes play in the regulation of these genes in ASD brain. Our goal is to better understand the molecular basis for the down-regulation of genes expressed in GABAergic neurons in ASD brains. We measured mRNA levels corresponding to selected GABAergic genes using qRT-PCR in RNA isolated from both ASD and CON groups. We determined the extent of binding of MECP2 and DNMT1 repressor proteins by chromatin immunoprecipitation (ChIP) assays. The amount of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) present in the promoters of the target genes was quantified by methyl DNA immunoprecipitation (MeDIP) and hydroxyl MeDIP (hMeDIP). We detected significant reductions in the mRNAs associated with RELN and GAD1 and significant increases in mRNAs encoding the Ten-eleven Translocation (TET) enzymes 1, 2, and 3. We also detected increased MECP2 and DNMT1 binding to the corresponding promoter regions of GAD1, RELN, and GAD2. Interestingly, there were decreased amounts of 5mC at both promoters and little change in 5hmC content in these same DNA fragments. Our data demonstrate that RELN, GAD1, and several other genes selectively expressed in GABAergic neurons, are down-regulated in post-mortem ASD FC. In addition, we observed increased DNMT1 and MECP2 binding at the corresponding promoters of these genes. The finding of increased MECP2 binding to the RELN, GAD1 and GAD2 promoters, with reduced amounts of 5mC and unchanged amounts of 5hmC present in these regions, suggests the possibility that DNMT1 interacts with and alters MECP2 binding properties to selected promoters. Comparisons between data obtained from the FC with CB studies showed some common themes between brain regions which are discussed.</p>',
'date' => '2017-02-14',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28229923',
'doi' => '',
'modified' => '2017-03-23 14:58:21',
'created' => '2017-03-23 14:58:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 59 => array(
'id' => '3126',
'name' => 'Intergenerational Transmission of Enhanced Seizure Susceptibility after Febrile Seizures',
'authors' => 'Wu D. et al.',
'description' => '<p>Environmental exposure early in development plays a role in susceptibility to disease in later life. Here, we demonstrate that prolonged febrile seizures induced by exposure of rat pups to a hyperthermic environment enhance seizure susceptibility not only in these hyperthermia-treated rats but also in their future offspring, even if the offspring never experience febrile seizures. This transgenerational transmission was intensity-dependent and was mainly from mothers to their offspring. The transmission was associated with DNA methylation. Thus, our study supports a “Lamarckian”-like mechanism of pathogenesis and the crucial role of epigenetic factors in neurological conditions.</p>',
'date' => '2017-02-08',
'pmid' => 'http://www.sciencedirect.com/science/article/pii/S2352396417300658',
'doi' => '',
'modified' => '2017-02-23 11:05:25',
'created' => '2017-02-23 11:05:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 60 => array(
'id' => '3125',
'name' => 'Pharmacological inhibition of DNA methyltransferase 1 promotes neuronal differentiation from rodent and human nasal olfactory stem/progenitor cell cultures',
'authors' => 'Franco I. et al.',
'description' => '<p>Nasal olfactory stem and neural progenitor cells (NOS/PCs) are considered possible tools for regenerative stem cell therapies in neurodegenerative diseases. Neurogenesis is a complex process regulated by extrinsic and intrinsic signals that include DNA-methylation and other chromatin modifications that could be experimentally manipulated in order to increase neuronal differentiation. The aim of the present study was the characterization of primary cultures and consecutive passages (P2-P10) of NOS/PCs isolated from male Swiss-Webster (mNOS/PCs) or healthy humans (hNOS/PCs). We evaluated and compared cellular morphology, proliferation rates and the expression pattern of pluripotency-associated markers and DNA methylation-associated gene expression in these cultures. Neuronal differentiation was induced by exposure to all-trans retinoic acid and forskolin for 7 days and evaluated by morphological analysis and immunofluorescence against neuronal markers MAP2, NSE and MAP1B. In response to the inductive cues mNOS/PCs expressed NSE (75.67%) and MAP2 (35.34%); whereas the majority of the hNOS/PCs were immunopositive to MAP1B. Treatment with procainamide, a specific inhibitor of DNA methyltransferase 1 (DNMT1), increases in the number of forskolin‘/retinoic acid-induced mature neuronal marker-expressing mNOS/PCs cells and enhances neurite development in hNOS/PCs. Our results indicate that mice and human nasal olfactory stem/progenitors cells share pluripotency-related gene expression suggesting that their application for stem cell therapy is worth pursuing and that DNA methylation inhibitors could be efficient tools to enhance neuronal differentiation from these cells.</p>',
'date' => '2017-02-01',
'pmid' => 'http://www.sciencedirect.com/science/article/pii/S0736574816303665',
'doi' => '',
'modified' => '2017-02-16 10:34:07',
'created' => '2017-02-16 10:34:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 61 => array(
'id' => '3119',
'name' => 'Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain',
'authors' => 'Amort T. et al.',
'description' => '<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec1">
<h3 xmlns="" class="Heading">Background</h3>
<p id="Par1" class="Para">Recent work has identified and mapped a range of posttranscriptional modifications in mRNA, including methylation of the N6 and N1 positions in adenine, pseudouridylation, and methylation of carbon 5 in cytosine (m5C). However, knowledge about the prevalence and transcriptome-wide distribution of m5C is still extremely limited; thus, studies in different cell types, tissues, and organisms are needed to gain insight into possible functions of this modification and implications for other regulatory processes.</p>
</div>
<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec2">
<h3 xmlns="" class="Heading">Results</h3>
<p id="Par2" class="Para">We have carried out an unbiased global analysis of m5C in total and nuclear poly(A) RNA of mouse embryonic stem cells and murine brain. We show that there are intriguing differences in these samples and cell compartments with respect to the degree of methylation, functional classification of methylated transcripts, and position bias within the transcript. Specifically, we observe a pronounced accumulation of m5C sites in the vicinity of the translational start codon, depletion in coding sequences, and mixed patterns of enrichment in the 3′ UTR. Degree and pattern of methylation distinguish transcripts modified in both embryonic stem cells and brain from those methylated in either one of the samples. We also analyze potential correlations between m5C and micro RNA target sites, binding sites of RNA binding proteins, and <em xmlns="" class="EmphasisTypeItalic">N</em>6-methyladenosine.</p>
</div>
<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec3">
<h3 xmlns="" class="Heading">Conclusion</h3>
<p id="Par3" class="Para">Our study presents the first comprehensive picture of cytosine methylation in the epitranscriptome of pluripotent and differentiated stages in the mouse. These data provide an invaluable resource for future studies of function and biological significance of m5C in mRNA in mammals.</p>
</div>',
'date' => '2017-01-05',
'pmid' => 'https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1139-1',
'doi' => '',
'modified' => '2017-02-14 17:20:08',
'created' => '2017-02-14 17:20:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 62 => array(
'id' => '3061',
'name' => 'Novel regional age-associated DNA methylation changes within human common disease-associated loci',
'authors' => 'Bell CG et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">Advancing age progressively impacts on risk and severity of chronic disease. It also modifies the epigenome, with changes in DNA methylation, due to both random drift and variation within specific functional loci.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">In a discovery set of 2238 peripheral-blood genome-wide DNA methylomes aged 19-82 years, we identify 71 age-associated differentially methylated regions within the linkage disequilibrium blocks of the single nucleotide polymorphisms from the NIH genome-wide association study catalogue. This included 52 novel regions, 29 within loci not covered by 450 k or 27 k Illumina array, and with enrichment for DNase-I Hypersensitivity sites across the full range of tissues. These age-associated differentially methylated regions also show marked enrichment for enhancers and poised promoters across multiple cell types. In a replication set of 2084 DNA methylomes, 95.7 % of the age-associated differentially methylated regions showed the same direction of ageing effect, with 80.3 % and 53.5 % replicated to p < 0.05 and p < 1.85 × 10<sup>-8</sup>, respectively.</abstracttext></p>
<h4>CONCLUSION:</h4>
<p><abstracttext label="CONCLUSION" nlmcategory="CONCLUSIONS">By analysing the functionally enriched disease and trait-associated regions of the human genome, we identify novel epigenetic ageing changes, which could be useful biomarkers or provide mechanistic insights into age-related common diseases.</abstracttext></p>
</div>',
'date' => '2016-09-26',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27663977',
'doi' => '',
'modified' => '2016-11-04 10:56:10',
'created' => '2016-11-02 09:54:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 63 => array(
'id' => '3007',
'name' => '5-hydroxymethylcytosine marks postmitotic neural cells in the adult and developing vertebrate central nervous system',
'authors' => 'Diotel N et al.',
'description' => '<p>The epigenetic mark 5-hydroxymethylcytosine (5hmC) is a cytosine modification that is abundant in the central nervous system of mammals and which results from 5-methylcytosine oxidation by TET enzymes. Such a mark is suggested to play key roles in the regulation of chromatin structure and gene expression. However, its precise functions still remain poorly understood and information about its distribution in non-mammalian species is still lacking. Here, the distribution of 5hmC was investigated in the brain of adult zebrafish, African claw frog, and mouse in a comparative manner. We show that zebrafish neurons are endowed with high levels of 5hmC, whereas quiescent or proliferative neural progenitors show low to undetectable levels of the modified cytosine. In the brain of larval and juvenile Xenopus, 5hmC is also detected in neurons, while ventricular proliferative cells do not display this epigenetic mark. Similarly, 5hmC is enriched in neurons compared to neural progenitors of the ventricular zone in the mouse developing cortex. Interestingly, 5hmC colocalized with the methylated DNA binding protein MeCP2 and with the active chromatin histone modification H3K4me2 in mouse neurons. Taken together, our results show an evolutionarily conserved cerebral distribution of 5hmC between fish and tetrapods and reinforce the idea that 5hmC fulfills major functions in the control of chromatin activity in vertebrate neurons.</p>',
'date' => '2016-07-14',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27414756',
'doi' => '',
'modified' => '2016-08-29 09:24:44',
'created' => '2016-08-29 09:24:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 64 => array(
'id' => '2992',
'name' => 'Regulation of the DNA Methylation Landscape in Human Somatic Cell Reprogramming by the miR-29 Family',
'authors' => 'Hysolli E et al.',
'description' => 'Reprogramming to pluripotency after overexpression of OCT4, SOX2, KLF4, and MYC is accompanied by global genomic and epigenomic changes. Histone modification and DNA methylation states in induced pluripotent stem cells (iPSCs) have been shown to be highly similar to embryonic stem cells (ESCs). However, epigenetic differences still exist between iPSCs and ESCs. In particular, aberrant DNA methylation states found in iPSCs are a major concern when using iPSCs in a clinical setting. Thus, it is critical to find factors that regulate DNA methylation states in reprogramming. Here, we found that the miR-29 family is an important epigenetic regulator during human somatic cell reprogramming. Our global DNA methylation and hydroxymethylation analysis shows that DNA demethylation is a major event mediated by miR-29a depletion during early reprogramming, and that iPSCs derived from miR-29a depletion are epigenetically closer to ESCs. Our findings uncover an important miRNA-based approach to generate clinically robust iPSCs.',
'date' => '2016-07-12',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27373925',
'doi' => '10.1016/j.stemcr.2016.05.014',
'modified' => '2016-08-23 09:57:29',
'created' => '2016-08-23 09:57:29',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 65 => array(
'id' => '3011',
'name' => 'Genome-Wide DNA Methylation in Mixed Ancestry Individuals with Diabetes and Prediabetes from South Africa',
'authors' => 'Matsha TE et al.',
'description' => '<p>Aims. To conduct a genome-wide DNA methylation in individuals with type 2 diabetes, individuals with prediabetes, and control mixed ancestry individuals from South Africa. Methods. We used peripheral blood to perform genome-wide DNA methylation analysis in 3 individuals with screen detected diabetes, 3 individuals with prediabetes, and 3 individuals with normoglycaemia from the Bellville South Community, Cape Town, South Africa, who were age-, gender-, body mass index-, and duration of residency-matched. Methylated DNA immunoprecipitation (MeDIP) was performed by Arraystar Inc. (Rockville, MD, USA). Results. Hypermethylated DMRs were 1160 (81.97%) and 124 (43.20%), respectively, in individuals with diabetes and prediabetes when both were compared to subjects with normoglycaemia. Our data shows that genes related to the immune system, signal transduction, glucose transport, and pancreas development have altered DNA methylation in subjects with prediabetes and diabetes. Pathway analysis based on the functional analysis mapping of genes to KEGG pathways suggested that the linoleic acid metabolism and arachidonic acid metabolism pathways are hypomethylated in prediabetes and diabetes. Conclusions. Our study suggests that epigenetic changes are likely to be an early process that occurs before the onset of overt diabetes. Detailed analysis of DMRs that shows gradual methylation differences from control versus prediabetes to prediabetes versus diabetes in a larger sample size is required to confirm these findings.</p>',
'date' => '2016-06-28',
'pmid' => 'http://www.hindawi.com/journals/ije/2016/3172093/',
'doi' => '',
'modified' => '2016-08-29 10:27:14',
'created' => '2016-08-29 10:27:14',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 66 => array(
'id' => '2954',
'name' => 'Dnmt2/Trdmt1 as Mediator of RNA Polymerase II Transcriptional Activity in Cardiac Growth',
'authors' => 'Ghanbarian H et al.',
'description' => '<p>Dnmt2/Trdmt1 is a methyltransferase, which has been shown to methylate tRNAs. Deficient mutants were reported to exhibit various, seemingly unrelated, defects in development and RNA-mediated epigenetic heredity. Here we report a role in a distinct developmental regulation effected by a noncoding RNA. We show that Dnmt2-deficiency in mice results in cardiac hypertrophy. Echocardiographic measurements revealed that cardiac function is preserved notwithstanding the increased dimensions of the organ due to cardiomyocyte enlargement. Mechanistically, activation of the P-TEFb complex, a critical step for cardiac growth, results from increased dissociation of the negatively regulating Rn7sk non-coding RNA component in Dnmt2-deficient cells. Our data suggest that Dnmt2 plays an unexpected role for regulation of cardiac growth by modulating activity of the P-TEFb complex.</p>',
'date' => '2016-06-06',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27270731',
'doi' => ' 10.1371/journal.pone.0156953',
'modified' => '2016-06-14 15:49:17',
'created' => '2016-06-14 15:49:17',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 67 => array(
'id' => '2941',
'name' => 'Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers',
'authors' => 'Li L et al.',
'description' => '<p>Promoter CpG methylation is a fundamental regulatory process of gene expression. TET proteins are active CpG demethylases converting 5-methylcytosine to 5-hydroxymethylcytosine, with loss of 5 hmC as an epigenetic hallmark of cancers, indicating critical roles of TET proteins in epigenetic tumorigenesis. Through analysis of tumor methylomes, we discovered <i>TET1</i> as a methylated target, and further confirmed its frequent downregulation/methylation in cell lines and primary tumors of multiple carcinomas and lymphomas, including nasopharyngeal, esophageal, gastric, colorectal, renal, breast and cervical carcinomas, as well as non-Hodgkin, Hodgkin and nasal natural killer/T-cell lymphomas, although all three <i>TET</i> family genes are ubiquitously expressed in normal tissues. Ectopic expression of TET1 catalytic domain suppressed colony formation and induced apoptosis of tumor cells of multiple tissue types, supporting its role as a broad <i>bona fide</i> tumor suppressor. Furthermore, TET1 catalytic domain possessed demethylase activity in cancer cells, being able to inhibit the CpG methylation of tumor suppressor gene (TSG) promoters and reactivate their expression, such as <i>SLIT2, ZNF382</i> and <i>HOXA9</i>. As only infrequent mutations of <i>TET1</i> have been reported, compared to <i>TET2</i>, epigenetic silencing therefore appears to be the dominant mechanism for <i>TET1</i> inactivation in cancers, which also forms a feedback loop of CpG methylation during tumorigenesis.</p>',
'date' => '2016-05-26',
'pmid' => 'http://www.nature.com/articles/srep26591',
'doi' => '10.1038/srep26591',
'modified' => '2016-06-06 09:47:31',
'created' => '2016-06-06 09:47:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 68 => array(
'id' => '2836',
'name' => 'Biochemical reconstitution of TET1–TDG–BER-dependent active DNA demethylation reveals a highly coordinated mechanism',
'authors' => 'Weber AR, Krawczyk C, Robertson AB, Kuśnierczyk A, Vågbø CB, Schuermann D, Klungland A, Schär P',
'description' => '<p>Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten–eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET–TDG–BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.nature.com/ncomms/2016/160302/ncomms10806/full/ncomms10806.html',
'doi' => '10.1038/ncomms10806',
'modified' => '2016-03-08 10:25:46',
'created' => '2016-03-08 10:25:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 69 => array(
'id' => '2837',
'name' => 'Hydroxymethylation of microRNA-365-3p Regulates Nociceptive Behaviors via Kcnh2',
'authors' => 'Pan Z, Zhang M, Ma T, Xue Z-Y, Li G-F, Hao L-Y, Zhu L-J, Li Y-Q, Ding H-L, Cao J-L',
'description' => '<p id="p-3">DNA 5-hydroxylmethylcytosine (5hmC) catalyzed by ten-eleven translocation methylcytosine dioxygenase (TET) occurs abundantly in neurons of mammals. However, the <em>in vivo</em> causal link between TET dysregulation and nociceptive modulation has not been established. Here, we found that spinal TET1 and TET3 were significantly increased in the model of formalin-induced acute inflammatory pain, which was accompanied with the augment of genome-wide 5hmC content in spinal cord. Knockdown of spinal TET1 or TET3 alleviated the formalin-induced nociceptive behavior and overexpression of spinal TET1 or TET3 in naive mice produced pain-like behavior as evidenced by decreased thermal pain threshold. Furthermore, we found that TET1 or TET3 regulated the nociceptive behavior by targeting microRNA-365-3p (miR-365-3p). Formalin increased 5hmC in the miR-365-3p promoter, which was inhibited by knockdown of TET1 or TET3 and mimicked by overexpression of TET1 or TET3 in naive mice. Nociceptive behavior induced by formalin or overexpression of spinal TET1 or TET3 could be prevented by downregulation of miR-365-3p, and mimicked by overexpression of spinal miR-365-3p. Finally, we demonstrated that a potassium channel, voltage-gated eag-related subfamily H member 2 (<em>Kcnh2</em>), validated as a target of miR-365-3p, played a critical role in nociceptive modulation by spinal TET or miR-365-3p. Together, we concluded that TET-mediated hydroxymethylation of miR-365-3p regulates nociceptive behavior via <em>Kcnh2</em>.</p>
<p id="p-4"><strong>SIGNIFICANCE STATEMENT</strong> Mounting evidence indicates that epigenetic modifications in the nociceptive pathway contribute to pain processes and analgesia response. Here, we found that the increase of 5hmC content mediated by TET1 or TET3 in miR-365-3p promoter in the spinal cord is involved in nociceptive modulation through targeting a potassium channel, <em>Kcnh2</em>. Our study reveals a new epigenetic mechanism underlying nociceptive information processing, which may be a novel target for development of antinociceptive drugs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.jneurosci.org/content/36/9/2769.short',
'doi' => '10.1523/JNEUROSCI.3474-15.2016',
'modified' => '2016-03-08 10:40:48',
'created' => '2016-03-08 10:40:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 70 => array(
'id' => '3034',
'name' => 'Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism',
'authors' => 'Weber AR et al.',
'description' => '<p>Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten-eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET-TDG-BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26932196',
'doi' => '',
'modified' => '2016-09-23 16:34:57',
'created' => '2016-09-23 16:34:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 71 => array(
'id' => '2833',
'name' => 'Genome-wide DNA methylation profile of developing deciduous tooth germ in miniature pigs',
'authors' => 'Su Y, Fan Z, Wu X, Li Y, Wang F, Zhang C, Wang J, Du J, Wang S',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND"><span class="highlight">DNA</span> <span class="highlight">methylation</span> is an important epigenetic modification critical to the regulation of gene expression during development. To date, little is known about the role of <span class="highlight">DNA</span> <span class="highlight">methylation</span> in <span class="highlight">tooth</span> development in large animal models. Thus, we carried out a comparative genomic analysis of <span class="highlight">genome-wide</span> <span class="highlight">DNA</span> <span class="highlight">methylation</span> profiles in E50 and E60 <span class="highlight">tooth</span> <span class="highlight">germ</span> from <span class="highlight">miniature</span> <span class="highlight">pigs</span> using methylated <span class="highlight">DNA</span> immunoprecipitation-sequencing (MeDIP-seq).</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We observed different <span class="highlight">DNA</span> <span class="highlight">methylation</span> patterns during the different developmental stages of pig <span class="highlight">tooth</span> <span class="highlight">germ</span>. A total of 2469 differentially methylated genes were identified. Functional analysis identified several signaling pathways and 104 genes that may be potential key regulators of pig <span class="highlight">tooth</span> development from E50 to E60.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">The present study provided a comprehensive analysis of the global <span class="highlight">DNA</span> <span class="highlight">methylation</span> pattern of <span class="highlight">tooth</span> <span class="highlight">germ</span> in <span class="highlight">miniature</span> <span class="highlight">pigs</span> and identified candidate genes that potentially regulate <span class="highlight">tooth</span> development from E50 to E60.</abstracttext></p>
</div>',
'date' => '2016-02-24',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26911717',
'doi' => '10.1186/s12864-016-2485-9',
'modified' => '2016-03-01 10:35:04',
'created' => '2016-03-01 10:35:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 72 => array(
'id' => '2834',
'name' => 'Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.',
'authors' => 'Rebollo R, Mager DL',
'description' => '<p>Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method used in our laboratory to detect, quantify, and compare mammalian endogenous retrovirus DNA methylation. More specifically we describe methylated DNA immunoprecipitation (MeDIP) followed by quantitative PCR.</p>',
'date' => '2016-02-20',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26895065',
'doi' => '10.1007/978-1-4939-3372-3_23',
'modified' => '2016-03-02 10:37:15',
'created' => '2016-03-02 10:22:16',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 73 => array(
'id' => '2815',
'name' => 'The dual specificity phosphatase 2 gene is hypermethylated in human cancer and regulated by epigenetic mechanisms',
'authors' => 'Tanja Haag, Antje M. Richter, Martin B. Schneider, Adriana P. Jiménez and Reinhard H. Dammann',
'description' => '<p><span>Dual specificity phosphatases are a class of tumor-associated proteins involved in the negative regulation of the MAP kinase pathway. Downregulation of the </span><em xmlns="" class="EmphasisTypeItalic">dual specificity phosphatase 2</em><span> (</span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span>) has been reported in cancer. Epigenetic silencing of tumor suppressor genes by abnormal promoter methylation is a frequent mechanism in oncogenesis. It has been shown that the epigenetic factor CTCF is involved in the regulation of tumor suppressor genes.</span></p>
<p><span>We analyzed the promoter hypermethylation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> in human cancer, including primary Merkel cell carcinoma by bisulfite restriction analysis and pyrosequencing. Moreover we analyzed the impact of a DNA methyltransferase inhibitor (5-Aza-dC) and CTCF on the epigenetic regulation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> by qRT-PCR, promoter assay, chromatin immuno-precipitation and methylation analysis.</span></span></p>
<p><span><span>Here we report a significant tumor-specific hypermethylation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> in primary Merkel cell carcinoma (</span><em xmlns="" class="EmphasisTypeItalic">p</em><span> = 0.05). An increase in methylation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> was also found in 17 out of 24 (71 %) cancer cell lines, including skin and lung cancer. Treatment of cancer cells with 5-Aza-dC induced </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression by its promoter demethylation, Additionally we observed that CTCF induces </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression in cell lines that exhibit silencing of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span>. This reactivation was accompanied by increased CTCF binding and demethylation of the </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> promoter.</span></span></span></p>
<p><span><span><span>Our data show that aberrant epigenetic inactivation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> occurs in carcinogenesis and that CTCF is involved in the epigenetic regulation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression.</span></span></span></span></p>',
'date' => '2016-02-01',
'pmid' => 'http://pubmed.gov/26833217',
'doi' => '10.1186/s12885-016-2087-6',
'modified' => '2016-03-09 16:34:51',
'created' => '2016-02-09 14:47:36',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 74 => array(
'id' => '2859',
'name' => 'Role of Growth Arrest and DNA Damage-Inducible, Beta in Alcohol-Drinking Behaviors',
'authors' => 'Gavin DP, Kusumo H, Zhang H, Guidotti A, Pandey SC',
'description' => '<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">The contribution of epigenetic factors, such as histone acetylation and DNA methylation, to the regulation of alcohol-drinking behavior has been increasingly recognized over the last several years. GADD45b is a protein demonstrated to be involved in DNA demethylation at neurotrophic factor gene promoters, including at brain-derived neurotrophic factor (Bdnf) which has been highly implicated in alcohol-drinking behavior.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">DNA methyltransferase-1 (Dnmt1), 3a, and 3b, and Gadd45a, b, and g mRNA were measured in the nucleus accumbens (NAc) and ventral tegmental areas of high ethanol (EtOH) consuming C57BL/6J (C57) and low alcohol consuming DBA/2J (DBA) mice using quantitative reverse transcriptase polymerase chain reaction (PCR). In the NAc, GADD45b protein was measured via immunohistochemistry and Bdnf9a mRNA using in situ PCR. Bdnf9a promoter histone H3 acetylated at lysines 9 and 14 (H3K9,K14ac) was measured using chromatin immunoprecipitation, and 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) using methylated DNA immunoprecipitation. Alcohol-drinking behavior was evaluated in Gadd45b haplodeficient (+/-) and null mice (-/-) utilizing drinking-in-the-dark (DID) and 2-bottle free-choice paradigms.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">C57 mice had lower levels of Gadd45b and g mRNA and GADD45b protein in the NAc relative to the DBA strain. C57 mice had lower NAc shell Bdnf9a mRNA levels, Bdnf9a promoter H3K9,K14ac, and higher Bdnf9a promoter 5HMC and 5MC. Acute EtOH increased GADD45b protein, Bdnf9a mRNA, and histone acetylation and decreased 5HMC in C57 mice. Gadd45b +/- mice displayed higher drinking behavior relative to wild-type littermates in both DID and 2-bottle free-choice paradigms.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">These data indicate the importance of the DNA demethylation pathway and its interactions with histone posttranslational modifications in alcohol-drinking behavior. Further, we suggest that lower DNA demethylation protein GADD45b levels may affect Bdnf expression possibly leading to altered alcohol-drinking behavior.</abstracttext></p>',
'date' => '2016-02-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26842245',
'doi' => ' 10.1111/acer.12965',
'modified' => '2016-03-15 16:37:22',
'created' => '2016-03-15 16:37:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 75 => array(
'id' => '2975',
'name' => 'Protocol for Methylated DNA Immunoprecipitation (MeDIP) Analysis',
'authors' => 'Karpova NN et al.',
'description' => '<p>DNA methylation is a fundamental epigenetic mechanism for silencing gene expression by either modifying chromatin structure to a repressive state or interfering with the transcription factors’ binding. DNA methylation primarily occurs at the position C5 of a cytosine ring mainly in the context of CpG dinucleotides. The modification can be recognized both in vivo and in vitro by the methyl-CpG binding proteins (MBPs) as well as in vitro by an antibody raised against 5-methylcytosine (5mC). This chapter describes different MBPs and introduces a standard methylated DNA immunoprecipitation (MeDIP) method, which is based on using the anti-5mC antibody to isolate methylated DNA fragments for subsequent locus-specific DNA methylation analysis. The MeDIP-generated DNA can be used as well for methylation profiling on a genome scale using array-based (MeDIP-chip) and high-throughput (MeDIP-seq) technologies.</p>',
'date' => '2016-02-01',
'pmid' => 'http://link.springer.com/protocol/10.1007/978-1-4939-2754-8_6',
'doi' => '10.1007/978-1-4939-2754-8_6',
'modified' => '2016-07-07 09:35:44',
'created' => '2016-07-07 09:35:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 76 => array(
'id' => '2844',
'name' => 'De novo DNA methylation drives 5hmC accumulation in mouse zygotes',
'authors' => 'Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, Koseki H, Sasaki H, Hajkova P',
'description' => '<p>Zygotic epigenetic reprogramming entails genome-wide DNA demethylation that is accompanied by Tet methylcytosine dioxygenase 3 (Tet3)-driven oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC; refs ,,,). Here we demonstrate using detailed immunofluorescence analysis and ultrasensitive LC-MS-based quantitative measurements that the initial loss of paternal 5mC does not require 5hmC formation. Small-molecule inhibition of Tet3 activity, as well as genetic ablation, impedes 5hmC accumulation in zygotes without affecting the early loss of paternal 5mC. Instead, 5hmC accumulation is dependent on the activity of zygotic Dnmt3a and Dnmt1, documenting a role for Tet3-driven hydroxylation in targeting de novo methylation activities present in the early embryo. Our data thus provide further insights into the dynamics of zygotic reprogramming, revealing an intricate interplay between DNA demethylation, de novo methylation and Tet3-driven hydroxylation.</p>',
'date' => '2016-01-11',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26751286',
'doi' => '10.1038/ncb3296',
'modified' => '2016-03-09 17:32:33',
'created' => '2016-03-09 17:32:33',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 77 => array(
'id' => '2860',
'name' => 'DNA methylation profiling: comparison of genome-wide sequencing methods and the Infinium Human Methylation 450 Bead Chip',
'authors' => 'Walker DL, Bhagwate AV, Baheti S, Smalley RL, Hilker CA, Sun Z, Cunningham JM',
'description' => '<div class="">
<h4>AIMS:</h4>
<p><abstracttext label="AIMS" nlmcategory="OBJECTIVE">To compare the performance of four sequence-based and one microarray methods for DNA methylation profiling.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">DNA from two cell lines were profiled by reduced representation bisulfite sequencing, methyl capture sequencing (SS-Meth Seq), NimbleGen SeqCapEpi CpGiant(Nimblegen MethSeq), methylated DNA immunoprecipitation (MeDIP) and the Human Methylation 450 Bead Chip (Meth450K).</abstracttext></p>
<h4>RESULTS & CONCLUSION:</h4>
<p><abstracttext label="RESULTS & CONCLUSION" nlmcategory="CONCLUSIONS">Despite differences in genome-wide coverage, high correlation and concordance were observed between different methods. Significant overlap of differentially methylated regions was identified between sequenced-based platforms. MeDIP provided the best coverage for the whole genome and gene body regions, while RRBS and Nimblegen MethSeq were superior for CpGs in CpG islands and promoters. Methylation analyses can be achieved by any of the five methods but understanding their differences may better address the research question being posed.</abstracttext></p>
</div>',
'date' => '2015-12-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26192535',
'doi' => '10.2217/EPI.15.64',
'modified' => '2016-03-16 11:06:05',
'created' => '2016-03-16 11:06:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 78 => array(
'id' => '2931',
'name' => 'Oxidative DNA damage in mouse sperm chromosomes: Size matters.',
'authors' => 'Kocer A et al.',
'description' => '<p>Normal embryo and foetal development as well as the health of the progeny are mostly dependent on gamete nuclear integrity. In the present study, in order to characterize more precisely oxidative DNA damage in mouse sperm we used two mouse models that display high levels of sperm oxidative DNA damage, a common alteration encountered both in in vivo and in vitro reproduction. Immunoprecipitation of oxidized sperm DNA coupled to deep sequencing showed that mouse chromosomes may be largely affected by oxidative alterations. We show that the vulnerability of chromosomes to oxidative attack inversely correlated with their size and was not linked to their GC richness. It was neither correlated with the chromosome content in persisting nucleosomes nor associated with methylated sequences. A strong correlation was found between oxidized sequences and sequences rich in short interspersed repeat elements (SINEs). Chromosome position in the sperm nucleus as revealed by fluorescent in situ hybridization appears to be a confounder. These data map for the first time fragile mouse sperm chromosomal regions when facing oxidative damage that may challenge the repair mechanisms of the oocyte post-fertilization.</p>',
'date' => '2015-12-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26510519',
'doi' => '10.1016/j.freeradbiomed.2015.10.419',
'modified' => '2016-05-19 10:18:24',
'created' => '2016-05-19 10:18:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 79 => array(
'id' => '2944',
'name' => 'Immunohistochemical Detection of Oxidized Forms of 5-Methylcytosine in Embryonic and Adult Brain Tissue',
'authors' => 'Abakir A et al.',
'description' => '<p>DNA methylation (5-methylcytosine, 5mC) is a major epigenetic modification of the eukaryotic genome associated with gene repression. Ten-eleven translocation proteins (Tet1/2/3) can oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Recent studies demonstrate that 5hmC is particularly enriched in neuronal cells and imply the involvement of this mark in transcriptional regulation taking place within the mammalian brain. Although a number of biochemical and antibody-based approaches have been successfully used to study the global content and genomic distributions of 5hmC in various contexts, most of these techniques do not provide any spatial information on the levels of this mark in different cell types. Here we describe a method of sensitive immunochemical detection of 5hmC/5fC/5caC in brain tissue based on the use of peroxidase-conjugated secondary antibodies and tyramide signal amplification. This technique can be instrumental in determining the relative enrichments of oxidized forms of 5mC in different brain cell types, effectively complementing other established approaches to investigate the functions of these marks in embryonic and adult brain.</p>',
'date' => '2015-09-02',
'pmid' => 'http://link.springer.com/protocol/10.1007%2F978-1-4939-2754-8_8',
'doi' => ' Print ISBN 978-1-4939-2753-1 Online ISBN 978-1-4939-2754-8 Series Title Neuromethods Series Volume 105 Series ISSN 0893-2336 Publisher Springer New York Copyright Holder Springer Science+Business Media New York Additional Lin',
'modified' => '2016-06-08 10:16:17',
'created' => '2016-06-08 10:16:17',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 80 => array(
'id' => '2847',
'name' => 'Gadd45b and N-methyl-D-aspartate induced DNA demethylation in postmitotic neurons.',
'authors' => 'Gavin DP, Kusumo H, Sharma RP, Guizzetti M, Guidotti A, Pandey SC.',
'description' => '<p><strong>AIM:</strong> In nondividing neurons examine the role of Gadd45b in active 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) removal at a gene promoter highly implicated in mental illnesses and cognition, Bdnf.</p>
<p><strong>MATERIALS & METHODS:</strong> Mouse primary cortical neuronal cultures with and without Gadd45b siRNA transfection were treated with N-methyl-d-aspartate (NMDA). Expression changes of genes reportedly involved in DNA demethylation, Bdnf mRNA and protein and 5MC and 5HMC at Bdnf promoters were measured.</p>
<p><strong>RESULTS:</strong> Gadd45b siRNA transfection in neurons abolishes the NMDA-induced increase in Bdnf IXa mRNA and reductions in 5MC and 5HMC at the Bdnf IXa promoter.</p>
<p><strong>CONCLUSION:</strong> These results contribute to our understanding of DNA demethylation mechanisms in neurons, and its role in regulating NMDA responsive genes implicated in mental illnesses.</p>',
'date' => '2015-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/26111030',
'doi' => '10.2217/epi.15.12',
'modified' => '2016-03-11 16:02:08',
'created' => '2016-03-11 15:47:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 81 => array(
'id' => '2879',
'name' => 'Active human nucleolar organizer regions are interspersed with inactive rDNA repeats in normal and tumor cells.',
'authors' => 'Zillner K, Komatsu J, Filarsky K, Kalepu R, Bensimon A, Németh A',
'description' => '<div class="">
<h4>AIM:</h4>
<p><abstracttext label="AIM" nlmcategory="OBJECTIVE">The synthesis of rRNA is a key determinant of normal and malignant cell growth and subject to epigenetic regulation. Yet, the epigenomic features of rDNA arrays clustered in nucleolar organizer regions are largely unknown. We set out to explore for the first time how DNA methylation is distributed on individual rDNA arrays.</abstracttext></p>
<h4>MATERIALS & METHODS:</h4>
<p><abstracttext label="MATERIALS & METHODS" nlmcategory="METHODS">Here we combined immunofluorescence detection of DNA modifications with fluorescence hybridization of single DNA fibers, metaphase immuno-FISH and methylation-sensitive restriction enzyme digestions followed by Southern blot.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We found clustering of both hypomethylated and hypermethylated repeat units and hypermethylation of noncanonical rDNA in IMR90 fibroblasts and HCT116 colorectal carcinoma cells. Surprisingly, we also found transitions between hypo- and hypermethylated rDNA repeat clusters on single DNA fibers.</abstracttext></p>
<h4>CONCLUSION:</h4>
<p><abstracttext label="CONCLUSION" nlmcategory="CONCLUSIONS">Collectively, our analyses revealed co-existence of different epialleles on individual nucleolar organizer regions and showed that epi-combing is a valuable approach to analyze epigenomic patterns of repetitive DNA.</abstracttext></p>
</div>',
'date' => '2015-06-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26077426',
'doi' => '10.2217/epi.14.93',
'modified' => '2016-04-05 09:44:29',
'created' => '2016-04-05 09:44:29',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 82 => array(
'id' => '2790',
'name' => 'Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency.',
'authors' => 'Chen H, Aksoy I, Gonnot F, Osteil P, Aubry M, Hamela C, Rognard C, Hochard A, Voisin S, Fontaine E, Mure M, Afanassieff M, Cleroux E, Guibert S, Chen J, Vallot C, Acloque H, Genthon C, Donnadieu C, De Vos J, Sanlaville D, Guérin JF, Weber M, Stanton LW, R',
'description' => 'Leukemia inhibitory factor (LIF)/STAT3 signalling is a hallmark of naive pluripotency in rodent pluripotent stem cells (PSCs), whereas fibroblast growth factor (FGF)-2 and activin/nodal signalling is required to sustain self-renewal of human PSCs in a condition referred to as the primed state. It is unknown why LIF/STAT3 signalling alone fails to sustain pluripotency in human PSCs. Here we show that the forced expression of the hormone-dependent STAT3-ER (ER, ligand-binding domain of the human oestrogen receptor) in combination with 2i/LIF and tamoxifen allows human PSCs to escape from the primed state and enter a state characterized by the activation of STAT3 target genes and long-term self-renewal in FGF2- and feeder-free conditions. These cells acquire growth properties, a gene expression profile and an epigenetic landscape closer to those described in mouse naive PSCs. Together, these results show that temporarily increasing STAT3 activity is sufficient to reprogramme human PSCs to naive-like pluripotent cells.',
'date' => '2015-05-13',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25968054',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 83 => array(
'id' => '2678',
'name' => 'Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells.',
'authors' => 'Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, Pop R, Reyon D, Tsai SQ, Mallard W, Joung JK, Rinn JL, Gnirke A, Meissner A',
'description' => 'DNA methylation is a key epigenetic modification involved in regulating gene expression and maintaining genomic integrity. Here we inactivated all three catalytically active DNA methyltransferases (DNMTs) in human embryonic stem cells (ESCs) using CRISPR/Cas9 genome editing to further investigate the roles and genomic targets of these enzymes. Disruption of DNMT3A or DNMT3B individually as well as of both enzymes in tandem results in viable, pluripotent cell lines with distinct effects on the DNA methylation landscape, as assessed by whole-genome bisulfite sequencing. Surprisingly, in contrast to findings in mouse, deletion of DNMT1 resulted in rapid cell death in human ESCs. To overcome this immediate lethality, we generated a doxycycline-responsive tTA-DNMT1* rescue line and readily obtained homozygous DNMT1-mutant lines. However, doxycycline-mediated repression of exogenous DNMT1* initiates rapid, global loss of DNA methylation, followed by extensive cell death. Our data provide a comprehensive characterization of DNMT-mutant ESCs, including single-base genome-wide maps of the targets of these enzymes.',
'date' => '2015-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25822089',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 84 => array(
'id' => '2477',
'name' => 'Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes.',
'authors' => 'Li L, Zhang Y, Fan Y, Sun K, Su X, Du Z, Tsao SW, Loh TK, Sun H, Chan AT, Zeng YX, Chan WY, Chan FK, Tao Q',
'description' => 'Aims: Nasopharyngeal carcinoma (NPC) is a common tumor consistently associated with Epstein-Barr virus infection and prevalent in South China, including Hong Kong, and southeast Asia. Current genomic sequencing studies found only rare mutations in NPC, indicating its critical epigenetic etiology, while no epigenome exists for NPC as yet. Materials & methods: We profiled the methylomes of NPC cell lines and primary tumors, together with normal nasopharyngeal epithelial cells, using methylated DNA immunoprecipitation (MeDIP). Results: We observed extensive, genome-wide methylation of cellular genes. Epigenetic disruption of Wnt, MAPK, TGF-β and Hedgehog signaling pathways was detected. Methylation of Wnt signaling regulators (SFRP1, 2, 4 and 5, DACT2, DKK2 and DKK3) was frequently detected in tumor and nasal swab samples from NPC patients. Functional studies showed that these genes are bona fide tumor-suppressor genes for NPC. Conclusion: The NPC methylome shows a special high-degree CpG methylation epigenotype, similar to the Epstein-Barr virus-infected gastric cancer, indicating a critical epigenetic etiology for NPC pathogenesis.',
'date' => '2015-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25479246',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 85 => array(
'id' => '2613',
'name' => 'CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.',
'authors' => 'Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC',
'description' => 'BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the underlying epigenetic signatures involved in its regulation. RESULTS: NOS3 and STAT3 mRNA levels were elevated in HUAEC of patients who suffered from placental insufficiency. 5-hydroxymethylcytosine, H3K9ac and Histone 2A (H2A).Zac at the NOS3 transcription start site directly correlated with NOS3 mRNA levels. Concomitantly, we observed entangled histone acetylation patterns and NOS3 response upon hypoxic conditions in vitro. Knock-down of either NOS3 or STAT3 by RNAi provided evidence for a functional NOS3/STAT3 relationship. Moreover, we recognized massive turnover of Stat3 at a discrete binding site in the NOS3 promoter. Interestingly, induced hyperacetylation resulted in short-termed increase of NOS3 mRNA followed by deferred decrease indicating that NOS3 expression could become self-attenuated by co-expressed intronic 27 nt-ncRNA. Reporter assay results and phylogenetic analyses enabled us to propose a novel model for STAT3-3'-UTR targeting by this 27-nt-ncRNA. CONCLUSIONS: An adverse intrauterine environment leads to adaptive changes of NOS3 expression. Apparently, a rapid NOS3 self-limiting response upon ectopic triggers co-exists with longer termed expression changes in response to placental insufficiency involving differential epigenetic signatures. Their persistence might contribute to impaired vascular endothelial response and consequently increase the risk of cardiovascular disease later in life.',
'date' => '2015-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25699114',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 86 => array(
'id' => '2437',
'name' => 'Acute Depletion Redefines the Division of Labor among DNA Methyltransferases in Methylating the Human Genome.',
'authors' => 'Tiedemann RL, Putiri EL, Lee JH, Hlady RA, Kashiwagi K, Ordog T, Zhang Z, Liu C, Choi JH, Robertson KD',
'description' => 'Global patterns of DNA methylation, mediated by the DNA methyltransferases (DNMTs), are disrupted in all cancers by mechanisms that remain largely unknown, hampering their development as therapeutic targets. Combinatorial acute depletion of all DNMTs in a pluripotent human tumor cell line, followed by epigenome and transcriptome analysis, revealed DNMT functions in fine detail. DNMT3B occupancy regulates methylation during differentiation, whereas an unexpected interplay was discovered in which DNMT1 and DNMT3B antithetically regulate methylation and hydroxymethylation in gene bodies, a finding confirmed in other cell types. DNMT3B mediated non-CpG methylation, whereas DNMT3L influenced the activity of DNMT3B toward non-CpG versus CpG site methylation. Altogether, these data reveal functional targets of each DNMT, suggesting that isoform selective inhibition would be therapeutically advantageous.',
'date' => '2014-11-20',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25453758',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 87 => array(
'id' => '2368',
'name' => 'A B-cell targeting virus disrupts potentially protective genomic methylation patterns in lymphoid tissue by increasing global 5-hydroxmethylcytosine levels',
'authors' => 'Ciccone NA, Mwangi W, Ruzov A, Smith LP, Butter C, Nair V',
'description' => 'The mechanisms by which viruses modulate the immune system include changes in host genomic methylation. 5-hydroxmethylcytosine (5hmC) is the catalytic product of the Tet (Ten-11 translocation) family of enzymes and may serve as an intermediate of DNA demethylation. Recent reports suggest that 5hmC may confer consequences on cellular events including the pathogenesis of disease; in order to explore this possibility further we investigated both 5-methylcytosine (5mC) and 5hmC levels in healthy and diseased chicken bursas of Fabricius. We discovered that embryonic B-cells have high 5mC content while 5hmC decreases during bursa development. We propose that a high 5mC level protects from the mutagenic activity of the B-cell antibody diversifying enzyme activation induced deaminase (AID). In support of this view, AID mRNA increases significantly within the developing bursa from embryonic to post hatch stages while mRNAs that encode Tet family members 1 and 2 reduce over the same period. Moreover, our data revealed that infectious bursal disease virus (IBDV) disrupts this genomic methylation pattern causing a global increase in 5hmC levels in a mechanism that may involve increased Tet 1 and 2 mRNAs. To our knowledge this is the first time that a viral infection has been observed to cause global increases in genomic 5hmC within infected host tissues, underlining a mechanism that may involve the induction of B-cell genomic instability and cell death to facilitate viral egress.',
'date' => '2014-10-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/25338704',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 88 => array(
'id' => '2350',
'name' => 'Spontaneous sleep-wake cycle and sleep deprivation differently induce Bdnf1, Bdnf4 and Bdnf9a DNA methylation and transcripts levels in the basal forebrain and frontal cortex in rats.',
'authors' => 'Ventskovska O, Porkka-Heiskanen T, Karpova NN',
'description' => 'Brain-derived neurotrophic factor (Bdnf) regulates neuronal plasticity, slow wave activity and sleep homeostasis. Environmental stimuli control Bdnf expression through epigenetic mechanisms, but there are no data on epigenetic regulation of Bdnf by sleep or sleep deprivation. Here we investigated whether 5-methylcytosine (5mC) DNA modification at Bdnf promoters p1, p4 and p9 influences Bdnf1, Bdnf4 and Bdnf9a expression during the normal inactive phase or after sleep deprivation (SD) (3, 6 and 12 h, end-times being ZT3, ZT6 and ZT12) in rats in two brain areas involved in sleep regulation, the basal forebrain and cortex. We found a daytime variation in cortical Bdnf expression: Bdnf1 expression was highest at ZT6 and Bdnf4 lowest at ZT12. Such variation was not observed in the basal forebrain. Also Bdnf p1 and p9 methylation levels differed only in the cortex, while Bdnf p4 methylation did not vary in either area. Factorial analysis revealed that sleep deprivation significantly induced Bdnf1 and Bdnf4 with the similar pattern for Bdnf9a in both basal forebrain and cortex; 12 h of sleep deprivation decreased 5mC levels at the cortical Bdnf p4 and p9. Regression analysis between the 5mC promoter levels and the corresponding Bdnf transcript expression revealed significant negative correlations for the basal forebrain Bdnf1 and cortical Bdnf9a transcripts in only non-deprived rats, while these correlations were lost after sleep deprivation. Our results suggest that Bdnf transcription during the light phase of undisturbed sleep-wake cycle but not after SD is regulated at least partially by brain site-specific DNA methylation.',
'date' => '2014-09-15',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25223586',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 89 => array(
'id' => '2109',
'name' => 'Transient accumulation of 5-carboxylcytosine indicates involvement of active demethylation in lineage specification of neural stem cells.',
'authors' => 'Wheldon LM, Abakir A, Ferjentsik Z, Dudnakova T, Strohbuecker S, Christie D, Dai N, Guan S, Foster JM, Corrêa IR, Loose M, Dixon JE, Sottile V, Johnson AD, Ruzov A',
'description' => '5-Methylcytosine (5mC) is an epigenetic modification involved in regulation of gene activity during differentiation. Tet dioxygenases oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). Both 5fC and 5caC can be excised from DNA by thymine-DNA glycosylase (TDG) followed by regeneration of unmodified cytosine via the base excision repair pathway. Despite evidence that this mechanism is operative in embryonic stem cells, the role of TDG-dependent demethylation in differentiation and development is currently unclear. Here, we demonstrate that widespread oxidation of 5hmC to 5caC occurs in postimplantation mouse embryos. We show that 5fC and 5caC are transiently accumulated during lineage specification of neural stem cells (NSCs) in culture and in vivo. Moreover, 5caC is enriched at the cell-type-specific promoters during differentiation of NSCs, and TDG knockdown leads to increased 5fC/5caC levels in differentiating NSCs. Our data suggest that active demethylation contributes to epigenetic reprogramming determining lineage specification in embryonic brain.',
'date' => '2014-06-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24882006',
'doi' => '',
'modified' => '2015-07-24 15:39:03',
'created' => '2015-07-24 15:39:03',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 90 => array(
'id' => '2448',
'name' => 'Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring.',
'authors' => 'Itzhak Y, Ergui I, Young JI',
'description' => 'The high rate of methamphetamine (METH) abuse among young adults and women of childbearing age makes it imperative to determine the long-term effects of METH exposure on the offspring. We hypothesized that parental METH exposure modulates offspring behavior by disrupting epigenetic programming of gene expression in the brain. To simulate the human pattern of drug use, male and female C57Bl/6J mice were exposed to escalating doses of METH or saline from adolescence through adulthood; following mating, females continue to receive drug or saline through gestational day 17. F1 METH male offspring showed enhanced response to cocaine-conditioned reward and hyperlocomotion. Both F1 METH male and female offspring had reduced response to conditioned fear. Cross-fostering experiments have shown that certain behavioral phenotypes were modulated by maternal care of either METH or saline dams. Analysis of offspring hippocampal DNA methylation showed differentially methylated regions as a result of both METH in utero exposure and maternal care. Our results suggest that behavioral phenotypes and epigenotypes of offspring that were exposed to METH in utero are vulnerable to (a) METH exposure during embryonic development, a period when wide epigenetic reprogramming occurs, and (b) postnatal maternal care.Molecular Psychiatry advance online publication, 18 February 2014; doi:10.1038/mp.2014.7.',
'date' => '2014-02-18',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24535458',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 91 => array(
'id' => '1843',
'name' => 'Alterations of epigenetic signatures in hepatocyte nuclear factor 4α deficient mouse liver determined by improved ChIP-qPCR and (h)MeDIP-qPCR assays.',
'authors' => 'Zhang Q, Lei X, Lu H',
'description' => 'Hepatocyte nuclear factor 4α (HNF4α) is a liver-enriched transcription factor essential for liver development and function. In hepatocytes, HNF4α regulates a large number of genes important for nutrient/xenobiotic metabolism and cell differentiation and proliferation. Currently, little is known about the epigenetic mechanism of gene regulation by HNF4α. In this study, the global and specific alterations at the selected gene loci of representative histone modifications and DNA methylations were investigated in Hnf4a-deficient female mouse livers using the improved MeDIP-, hMeDIP- and ChIP-qPCR assay. Hnf4a deficiency significantly increased hepatic total IPed DNA fragments for histone H3 lysine-4 dimethylation (H3K4me2), H3K4me3, H3K9me2, H3K27me3 and H3K4 acetylation, but not for H3K9me3, 5-methylcytosine,or 5-hydroxymethylcytosine. At specific gene loci, the relative enrichments of histone and DNA modifications were changed to different degree in Hnf4a-deficient mouse liver. Among the epigenetic signatures investigated, changes in H3K4me3 correlated the best with mRNA expression. Additionally, Hnf4a-deficient livers had increased mRNA expression of histone H1.2 and H3.3 as well as epigenetic modifiers Dnmt1, Tet3, Setd7, Kmt2c, Ehmt2, and Ezh2. In conclusion, the present study provides convenient improved (h)MeDIP- and ChIP-qPCR assays for epigenetic study. Hnf4a deficiency in young-adult mouse liver markedly alters histone methylation and acetylation, with fewer effects on DNA methylation and 5-hydroxymethylation. The underlying mechanism may be the induction of epigenetic enzymes responsible for the addition/removal of the epigenetic signatures, and/or the loss of HNF4α per se as a key coordinator for epigenetic modifiers.',
'date' => '2014-01-10',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24427299',
'doi' => '',
'modified' => '2015-07-24 15:39:01',
'created' => '2015-07-24 15:39:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 92 => array(
'id' => '1773',
'name' => 'Peroxisome proliferator-activated receptor γ regulates genes involved in insulin/insulin-like growth factor signaling and lipid metabolism during adipogenesis through functionally distinct enhancer classes.',
'authors' => 'Oger F, Dubois-Chevalier J, Gheeraert C, Avner S, Durand E, Froguel P, Salbert G, Staels B, Lefebvre P, Eeckhoute J',
'description' => 'The nuclear receptor peroxisome proliferator-activated receptor (PPAR) is a transcription factor whose expression is induced during adipogenesis and that is required for the acquisition and control of mature adipocyte functions. Indeed, PPAR induces the expression of genes involved in lipid synthesis and storage through enhancers activated during adipocyte differentiation. Here, we show that PPAR also binds to enhancers already active in preadipocytes as evidenced by an active chromatin state including lower DNA methylation levels despite higher CpG content. These constitutive enhancers are linked to genes involved in the insulin/insulin-like growth factor signaling pathway that are transcriptionally induced during adipogenesis but to a lower extent than lipid metabolism genes, because of stronger basal expression levels in preadipocytes. This is consistent with the sequential involvement of hormonal sensitivity and lipid handling during adipocyte maturation and correlates with the chromatin structure dynamics at constitutive and activated enhancers. Interestingly, constitutive enhancers are evolutionary conserved and can be activated in other tissues, in contrast to enhancers controlling lipid handling genes whose activation is more restricted to adipocytes. Thus, PPAR utilizes both broadly active and cell type-specific enhancers to modulate the dynamic range of activation of genes involved in the adipogenic process.',
'date' => '2014-01-10',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24288131',
'doi' => '',
'modified' => '2015-07-24 15:39:01',
'created' => '2015-07-24 15:39:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 93 => array(
'id' => '1572',
'name' => 'Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model.',
'authors' => 'Yafune A, Kawai M, Itahashi M, Kimura M, Nakane F, Mitsumori K, Shibutani M',
'description' => 'Disruptive epigenetic gene control has been shown to be involved in carcinogenesis. To identify key molecules in piperonyl butoxide (PBO)-induced hepatocarcinogenesis, we searched hypermethylated genes using CpG island (CGI) microarrays in non-neoplastic liver cells as a source of proliferative lesions at 25 weeks after tumor promotion with PBO using mice. We further performed methylation-specific polymerase chain reaction (PCR), real-time reverse transcription PCR, and immunohistochemical analysis in PBO-promoted liver tissues. Ebp4.1, Wdr6 and Cmtm6 increased methylation levels in the promoter region by PBO promotion, although Cmtm6 levels were statistically non-significant. These results suggest that PBO promotion may cause altered epigenetic gene regulation in non-neoplastic liver cells surrounding proliferative lesions to allow the facilitation of hepatocarcinogenesis. Both Wdr6 and Cmtm6 showed decreased expression in non-neoplastic liver cells in contrast to positive immunoreactivity in the majority of proliferative lesions produced by PBO promotion. These results suggest that both Wdr6 and Cmtm6 were spared from epigenetic gene modification in proliferative lesions by PBO promotion in contrast to the hypermethylation-mediated downregulation in surrounding liver cells. Considering the effective detection of proliferative lesions, these molecules could be used as detection markers of hepatocellular proliferative lesions and played an important role in hepatocarcinogenesis.',
'date' => '2013-10-09',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23968726',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 94 => array(
'id' => '1569',
'name' => 'Genome-wide screening identifies Plasmodium chabaudi-induced modifications of DNA methylation status of Tlr1 and Tlr6 gene promoters in liver, but not spleen, of female C57BL/6 mice.',
'authors' => 'Al-Quraishy S, Dkhil MA, Abdel-Baki AA, Delic D, Santourlidis S, Wunderlich F',
'description' => '<p>Epigenetic reprogramming of host genes via DNA methylation is increasingly recognized as critical for the outcome of diverse infectious diseases, but information for malaria is not yet available. Here, we investigate the effect of blood-stage malaria of Plasmodium chabaudi on the DNA methylation status of host gene promoters on a genome-wide scale using methylated DNA immunoprecipitation and Nimblegen microarrays containing 2,000 bp oligonucleotide features that were split into -1,500 to -500 bp Ups promoters and -500 to +500 bp Cor promoters, relative to the transcription site, for evaluation of differential DNA methylation. Gene expression was analyzed by Agilent and Affymetrix microarray technology. Challenging of female C57BL/6 mice with 10(6) P. chabaudi-infected erythrocytes resulted in a self-healing outcome of infections with peak parasitemia on day 8 p.i. These infections induced organ-specific modifications of DNA methylation of gene promoters. Among the 17,354 features on Nimblegen arrays, only seven gene promoters were identified to be hypermethylated in the spleen, whereas the liver exhibited 109 hyper- and 67 hypomethylated promoters at peak parasitemia in comparison with non-infected mice. Among the identified genes with differentially methylated Cor-promoters, only the 7 genes Pigr, Ncf1, Klkb1, Emr1, Ndufb11, and Tlr6 in the liver and Apol6 in the spleen were detected to have significantly changed their expression. Remarkably, the Cor promoter of the toll-like receptor Tlr6 became hypomethylated and Tlr6 expression increased by 3.4-fold during infection. Concomitantly, the Ups promoter of the Tlr1 was hypermethylated, but Tlr1 expression also increased by 11.3-fold. TLR6 and TLR1 are known as auxillary receptors to form heterodimers with TLR2 in plasma membranes of macrophages, which recognize different pathogen-associated molecular patterns (PAMPs), as, e.g., intact 3-acyl and sn-2-lyso-acyl glycosylphosphatidylinositols of P. falciparum, respectively. Our data suggest therefore that malaria-induced epigenetic fine-tuning of Tlr6 and Tlr1 through DNA methylation of their gene promoters in the liver is critically important for initial recognition of PAMPs and, thus, for the final self-healing outcome of blood-stage infections with P. chabaudi malaria.</p>',
'date' => '2013-08-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23949311',
'doi' => '',
'modified' => '2017-10-10 10:37:58',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 95 => array(
'id' => '1466',
'name' => 'Characterization of the DNA methylome and its interindividual variation in human peripheral blood monocytes.',
'authors' => 'Shen H, Qiu C, Li J, Tian Q, Deng HW',
'description' => 'AIM: Peripheral blood monocytes (PBMs) play multiple and critical roles in the immune response, and abnormalities in PBMs have been linked to a variety of human disorders. However, the DNA methylation landscape in PBMs is largely unknown. In this study, we characterized epigenome-wide DNA methylation profiles in purified PBMs. MATERIALS & METHODS: PBMs were isolated from freshly collected peripheral blood from 18 unrelated healthy postmenopausal Caucasian females. Epigenome-wide DNA methylation profiles (the methylome) were characterized by using methylated DNA immunoprecipitation combined with high-throughput sequencing. RESULTS: Distinct patterns were revealed at different genomic features. For instance, promoters were commonly (∼58%) found to be unmethylated; whereas protein coding regions were largely (∼84%) methylated. Although CpG-rich and -poor promoters showed distinct methylation patterns, interestingly, a negative correlation between promoter methylation levels and gene transcription levels was consistently observed across promoters with high to low CpG densities. Importantly, we observed substantial interindividual variation in DNA methylation across the individual PBM methylomes and the pattern of this interindividual variation varied between different genomic features, with highly variable regions enriched for repetitive DNA elements. Furthermore, we observed a modest but significant excess (p < 2.2 × 10(-16)) of genes showing a negative correlation between interindividual promoter methylation and transcription levels. These significant genes were enriched in biological processes that are closely related to PBM functions, suggesting that alteration in DNA methylation is likely to be an important mechanism contributing to the interindividual variation in PBM function, and PBM-related phenotypic and disease-susceptibility variation in humans. CONCLUSION: This study represents a comprehensive analysis of the human PBM methylome and its interindividual variation. Our data provide a valuable resource for future epigenomic and multiomic studies, exploring biological and disease-related regulatory mechanisms in PBMs.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23750642',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 96 => array(
'id' => '1463',
'name' => 'Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation.',
'authors' => 'Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD',
'description' => 'Histone post-translational modifications regulate chromatin structure and function largely through interactions with effector proteins that often contain multiple histone-binding domains. While significant progress has been made characterizing individual effector domains, the role of paired domains and how they function in a combinatorial fashion within chromatin are poorly defined. Here we show that the linked tandem Tudor and plant homeodomain (PHD) of UHRF1 (ubiquitin-like PHD and RING finger domain-containing protein 1) operates as a functional unit in cells, providing a defined combinatorial readout of a heterochromatin signature within a single histone H3 tail that is essential for UHRF1-directed epigenetic inheritance of DNA methylation. These findings provide critical support for the "histone code" hypothesis, demonstrating that multivalent histone engagement plays a key role in driving a fundamental downstream biological event in chromatin.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23752590',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 97 => array(
'id' => '1449',
'name' => 'Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs.',
'authors' => 'Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R',
'description' => 'The presence of 5-methylcytidine (m(5)C) in tRNA and rRNA molecules of a wide variety of organisms was first observed more than 40 years ago. However, detection of this modification was limited to specific, abundant, RNA species, due to the usage of low-throughput methods. To obtain a high resolution, systematic, and comprehensive transcriptome-wide overview of m(5)C across the three domains of life, we used bisulfite treatment on total RNA from both gram positive (B. subtilis) and gram negative (E. coli) bacteria, an archaeon (S. solfataricus) and a eukaryote (S. cerevisiae), followed by massively parallel sequencing. We were able to recover most previously documented m(5)C sites on rRNA in the four organisms, and identified several novel sites in yeast and archaeal rRNAs. Our analyses also allowed quantification of methylated m(5)C positions in 64 tRNAs in yeast and archaea, revealing stoichiometric differences between the methylation patterns of these organisms. Molecules of tRNAs in which m(5)C was absent were also discovered. Intriguingly, we detected m(5)C sites within archaeal mRNAs, and identified a consensus motif of AUCGANGU that directs methylation in S. solfataricus. Our results, which were validated using m(5)C-specific RNA immunoprecipitation, provide the first evidence for mRNA modifications in archaea, suggesting that this mode of post-transcriptional regulation extends beyond the eukaryotic domain.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23825970',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 98 => array(
'id' => '1403',
'name' => 'Methyl donor supplementation blocks the adverse effects of maternal high fat diet on offspring physiology.',
'authors' => 'Carlin J, George R, Reyes TM',
'description' => 'Maternal consumption of a high fat diet during pregnancy increases the offspring risk for obesity. Using a mouse model, we have previously shown that maternal consumption of a high fat (60%) diet leads to global and gene specific decreases in DNA methylation in the brain of the offspring. The present experiments were designed to attempt to reverse this DNA hypomethylation through supplementation of the maternal diet with methyl donors, and to determine whether methyl donor supplementation could block or attenuate phenotypes associated with maternal consumption of a HF diet. Metabolic and behavioral (fat preference) outcomes were assessed in male and female adult offspring. Expression of the mu-opioid receptor and dopamine transporter mRNA, as well as global DNA methylation were measured in the brain. Supplementation of the maternal diet with methyl donors attenuated the development of some of the adverse effects seen in offspring from dams fed a high fat diet; including weight gain, increased fat preference (males), changes in CNS gene expression and global hypomethylation in the prefrontal cortex. Notable sex differences were observed. These findings identify the importance of balanced methylation status during pregnancy, particularly in the context of a maternal high fat diet, for optimal offspring outcome.',
'date' => '2013-05-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23658839',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 99 => array(
'id' => '1311',
'name' => 'Naive pluripotency is associated with global DNA hypomethylation.',
'authors' => 'Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, Surani MA, Hajkova P',
'description' => 'Naive pluripotent embryonic stem cells (ESCs) and embryonic germ cells (EGCs) are derived from the preimplantation epiblast and primordial germ cells (PGCs), respectively. We investigated whether differences exist between ESCs and EGCs, in view of their distinct developmental origins. PGCs are programmed to undergo global DNA demethylation; however, we find that EGCs and ESCs exhibit equivalent global DNA methylation levels. Inhibition of MEK and Gsk3b by 2i conditions leads to pronounced reduction in DNA methylation in both cell types. This is driven by Prdm14 and is associated with downregulation of Dnmt3a and Dnmt3b. However, genomic imprints are maintained in 2i, and we report derivation of EGCs with intact genomic imprints. Collectively, our findings establish that culture in 2i instills a naive pluripotent state with a distinctive epigenetic configuration that parallels molecular features observed in both the preimplantation epiblast and nascent PGCs.',
'date' => '2013-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23416945',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 100 => array(
'id' => '1290',
'name' => 'DNA methylation analysis in the intestinal epithelium-effect of cell separation on gene expression and methylation profile.',
'authors' => 'Jenke AC, Postberg J, Raine T, Nayak KM, Molitor M, Wirth S, Kaser A, Parkes M, Heuschkel RB, Orth V, Zilbauer M',
'description' => 'BACKGROUND: Epigenetic signatures are highly cell type specific. Separation of distinct cell populations is therefore desirable for all epigenetic studies. However, to date little information is available on whether separation protocols might influence epigenetic and/or gene expression signatures and hence might be less beneficial. We investigated the influence of two frequently used protocols to isolate intestinal epithelium cells (IECs) from 6 healthy individuals. MATERIALS AND METHODS: Epithelial cells were isolated from small bowel (i.e. terminal ileum) biopsies using EDTA/DTT and enzymatic release followed by magnetic bead sorting via EPCAM labeled microbeads. Effects on gene/mRNA expression were analyzed using a real time PCR based expression array. DNA methylation was assessed by pyrosequencing of bisulfite converted DNA and methylated DNA immunoprecipitation (MeDIP). RESULTS: While cell purity was >95% using both cell separation approaches, gene expression analysis revealed significantly higher mRNA levels of several inflammatory genes in EDTA/DTT when compared to enzymatically released cells. In contrast, DNA methylation of selected genes was less variable and only revealed subtle differences. Comparison of DNA methylation of the epithelial cell marker EPCAM in unseparated whole biopsy samples with separated epithelium (i.e. EPCAM positive and negative fraction) demonstrated significant differences in DNA methylation between all three tissue fractions indicating cell type specific methylation patterns can be masked in unseparated tissue samples. CONCLUSIONS: Taken together, our data highlight the importance of considering the potential effect of cell separation on gene expression as well as DNA methylation signatures. The decision to separate tissue samples will therefore depend on study design and specific separation protocols.',
'date' => '2013-02-08',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23409010',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 101 => array(
'id' => '1065',
'name' => 'Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation.',
'authors' => 'Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD',
'description' => 'A fundamental challenge in mammalian biology has been the elucidation of mechanisms linking DNA methylation and histone post-translational modifications. Human UHRF1 (ubiquitin-like PHD and RING finger domain-containing 1) has multiple domains that bind chromatin, and it is implicated genetically in the maintenance of DNA methylation. However, molecular mechanisms underlying DNA methylation regulation by UHRF1 are poorly defined. Here we show that UHRF1 association with methylated histone H3 Lys9 (H3K9) is required for DNA methylation maintenance. We further show that UHRF1 association with H3K9 methylation is insensitive to adjacent H3 S10 phosphorylation-a known mitotic 'phospho-methyl switch'. Notably, we demonstrate that UHRF1 mitotic chromatin association is necessary for DNA methylation maintenance through regulation of the stability of DNA methyltransferase-1. Collectively, our results define a previously unknown link between H3K9 methylation and the faithful epigenetic inheritance of DNA methylation, establishing a notable mitotic role for UHRF1 in this process.',
'date' => '2012-09-30',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23022729',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 102 => array(
'id' => '960',
'name' => 'Histone acetylation and DNA demethylation of T-cells result in an anaplastic large cell lymphoma-like phenotype.',
'authors' => 'Joosten M, Seitz V, Zimmermann K, Sommerfeld A, Berg E, Lenze D, Leser U, Stein H, Hummel M',
'description' => 'Background. A characteristic feature of anaplastic large cell lymphoma is the significant repression of the T-cell expression program despite its T-cell origin. The reasons for this down-regulation of T-cell phenotype are still unknown. Design and Methods. To elucidate whether epigenetic mechanisms are responsible for the loss of the T-cell phenotype, we treated anaplastic large cell lymphoma and T-cell lymphoma/leukemia cell lines (n=4, each) with epigenetic modifiers to evoke DNA demethylation and histone acetylation. Global gene expression data from treated and untreated cell lines were generated and selected differentially expressed genes were evaluated by real-time RT-PCR and Western Blot analysis. Additionally, histone H3 lysine 27 trimethylation was analyzed by chromatin immunoprecipitation. Results. Combined DNA demethylation and histone acetylation of anaplastic large cell lymphoma cells was not able to reconstitute their T-cell phenotype. Instead, the same treatment induced in T-cells (i) an up-regulation of anaplastic large cell lymphoma-characteristic genes (e.g. ID2, LGALS1, c-JUN) and (ii) an almost complete extinction of their T-cell phenotype including CD3, LCK and ZAP70. In addition, a suppressive trimethylation of histone H3 lysine 27 of important T-cell transcription factor genes (GATA3, LEF1, TCF1) was present in anaplastic large cell lymphoma cells, which is in line with their absence in primary tumour specimens as demonstrated by immunohistochemistry. Conclusions. Our data suggest that epigenetically activated suppressors (e.g. ID2) contribute to the down-regulation of the T-cell expression program in anaplastic large cell lymphoma, which is maintained by trimethylation of histone H3 lysine 27.',
'date' => '2012-08-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22899583',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 103 => array(
'id' => '389',
'name' => 'Growth Arrest and DNA-Damage-Inducible, Beta (GADD45b)-Mediated DNA Demethylation in Major Psychosis.',
'authors' => 'Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A',
'description' => 'Aberrant neocortical DNA methylation has been suggested to be a pathophysiological contributor to psychotic disorders. Recently, a growth arrest and DNA-damage-inducible, beta (GADD45b) protein-coordinated DNA demethylation pathway, utilizing cytidine deaminases and thymidine glycosylases, has been identified in the brain. We measured expression of several members of this pathway in parietal cortical samples from the Stanley Foundation Neuropathology Consortium (SFNC) cohort. We find an increase in GADD45b mRNA and protein in patients with psychosis. In immunohistochemistry experiments using samples from the Harvard Brain Tissue Resource Center, we report an increased number of GADD45b-stained cells in prefrontal cortical layers II, III, and V in psychotic patients. Brain-derived neurotrophic factor IX (BDNF IXabcd) was selected as a readout gene to determine the effects of GADD45b expression and promoter binding. We find that there is less GADD45b binding to the BDNF IXabcd promoter in psychotic subjects. Further, there is reduced BDNF IXabcd mRNA expression, and an increase in 5-methylcytosine and 5-hydroxymethylcytosine at its promoter. On the basis of these results, we conclude that GADD45b may be increased in psychosis compensatory to its inability to access gene promoter regions.',
'date' => '2012-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22048458',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 104 => array(
'id' => '409',
'name' => 'Epigenetic silencing mediated through activated PI3K/AKT signaling in breast cancer.',
'authors' => 'Zuo T, Liu TM, Lan X, Weng YI, Shen R, Gu F, Huang YW, Liyanarachchi S, Deatherage DE, Hsu PY, Taslim C, Ramaswamy B, Shapiro CL, Lin HJ, Cheng AS, Jin VX, Huang TH',
'description' => 'Trimethylation of histone 3 lysine 27 (H3K27me3) is a critical epigenetic mark for the maintenance of gene silencing. Additional accumulation of DNA methylation in target loci is thought to cooperatively support this epigenetic silencing during tumorigenesis. However, molecular mechanisms underlying the complex interplay between the two marks remain to be explored. Here we show that activation of PI3K/AKT signaling can be a trigger of this epigenetic processing at many downstream target genes. We also find that DNA methylation can be acquired at the same loci in cancer cells, thereby reinforcing permanent repression in those losing the H3K27me3 mark. Because of a link between PI3K/AKT signaling and epigenetic alterations, we conducted epigenetic therapies in conjunction with the signaling-targeted treatment. These combined treatments synergistically relieve gene silencing and suppress cancer cell growth in vitro and in xenografts. The new finding has important implications for improving targeted cancer therapies in the future.',
'date' => '2011-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/21216892',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 105 => array(
'id' => '412',
'name' => 'Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping.',
'authors' => 'Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, Weng YI, Deatherage DE, Chen Z, Pereira JS, Lopez R, Russo J, Wang Q, Lamartiniere CA, Nephew KP, Huang TH',
'description' => 'The current concept of epigenetic repression is based on one repressor unit corresponding to one silent gene. This notion, however, cannot adequately explain concurrent silencing of multiple loci observed in large chromosome regions. The long-range epigenetic silencing (LRES) can be a frequent occurrence throughout the human genome. To comprehensively characterize the influence of estrogen signaling on LRES, we analyzed transcriptome, methylome, and estrogen receptor alpha (ESR1)-binding datasets from normal breast epithelia and breast cancer cells. This "omics" approach uncovered 11 large repressive zones (range, 0.35 approximately 5.98 megabases), including a 14-gene cluster located on 16p11.2. In normal cells, estrogen signaling induced transient formation of multiple DNA loops in the 16p11.2 region by bringing 14 distant loci to focal ESR1-docking sites for coordinate repression. However, the plasticity of this free DNA movement was reduced in breast cancer cells. Together with the acquisition of DNA methylation and repressive chromatin modifications at the 16p11.2 loci, an inflexible DNA scaffold may be a novel determinant used by breast cancer cells to reinforce estrogen-mediated repression.',
'date' => '2010-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/20442245',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 106 => array(
'id' => '976',
'name' => 'Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells.',
'authors' => 'Borgel J, Guibert S, Weber M.',
'description' => 'Methylated DNA immunoprecipitation (MeDIP) is an immunocapturing approach for unbiased enrichment of DNA that is methylated on cytosines. The principle is that genomic DNA is randomly sheared by sonication and immunoprecipitated with an antibody that specifically recognizes 5-methylcytidine (5mC), which can be combined with PCR or high-throughput analysis (microarrays, deep sequencing). The MeDIP technique has been originally used to generate DNA methylation profiles on a genome scale in mammals and plants. Here we provide an optimized version of the MeDIP protocol suitable for low amounts of DNA, which can be used to study DNA methylation in cellular populations available in small quantities.',
'date' => '0000-00-00',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22907495',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '120',
'name' => '5-mC Antibody - clone 33D3 SDS US en',
'language' => 'en',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-US-en-GHS_2_0.pdf',
'countries' => 'US',
'modified' => '2020-03-13 15:47:06',
'created' => '2020-03-13 15:47:06',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '118',
'name' => '5-mC Antibody - clone 33D3 SDS GB en',
'language' => 'en',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-GB-en-GHS_2_0.pdf',
'countries' => 'GB',
'modified' => '2020-03-13 15:44:22',
'created' => '2020-03-13 15:44:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '116',
'name' => '5-mC Antibody - clone 33D3 SDS ES es',
'language' => 'es',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-ES-es-GHS_2_0.pdf',
'countries' => 'ES',
'modified' => '2020-03-13 15:43:01',
'created' => '2020-03-13 15:43:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '115',
'name' => '5-mC Antibody - clone 33D3 SDS DE de',
'language' => 'de',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-DE-de-GHS_2_0.pdf',
'countries' => 'DE',
'modified' => '2020-03-13 15:42:22',
'created' => '2020-03-13 15:42:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '119',
'name' => '5-mC Antibody - clone 33D3 SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-JP-ja-GHS_4_0.pdf',
'countries' => 'JP',
'modified' => '2020-03-13 15:44:56',
'created' => '2020-03-13 15:44:56',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '114',
'name' => '5-mC Antibody - clone 33D3 SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-03-13 15:41:00',
'created' => '2020-03-13 15:41:00',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '113',
'name' => '5-mC Antibody - clone 33D3 SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-BE-fr-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-03-13 15:40:12',
'created' => '2020-03-13 15:40:12',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '117',
'name' => '5-mC Antibody - clone 33D3 SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-03-13 15:43:48',
'created' => '2020-03-13 15:43:48',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = true
$other_formats = array(
(int) 0 => array(
'id' => '1980',
'antibody_id' => '630',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>Monoclonal antibody raised in mouse against </span><b>5-mC</b><span><span> </span>(</span><b>5-methylcytosine</b><span>) conjugated to ovalbumine (</span><b>33D3 clone</b><span>).</span></p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15200081-100',
'old_catalog_number' => 'MAb-081-100',
'sf_code' => 'C15200081-D001-000526',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '505',
'price_USD' => '575',
'price_GBP' => '450',
'price_JPY' => '79110',
'price_CNY' => '0',
'price_AUD' => '1438',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'October 27, 2020',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081) | Diagenode',
'meta_keywords' => '5-methylcytosine (5-mC),monoclonal antibody,Methylated DNA Immunoprecipitation',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2023-05-17 10:08:33',
'created' => '2015-06-29 14:08:20'
),
(int) 1 => array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
)
$pro = array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010034</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1885" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1885" id="CartAdd/1885Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1885" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto hMeDIP kit x16 (monoclonal mouse antibody)</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns" data-reveal-id="cartModal-1885" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto hMeDIP kit x16 (monoclonal mouse antibody)</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/magmedip-kit-x48-48-rxns"><img src="/img/product/kits/C02010021-magmedip-qpcr.jpg" alt="MagMeDIP qPCR Kit box" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010021</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1880" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1880" id="CartAdd/1880Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1880" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MagMeDIP Kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MagMeDIP Kit',
'C02010021',
'750',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MagMeDIP Kit',
'C02010021',
'750',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="magmedip-kit-x48-48-rxns" data-reveal-id="cartModal-1880" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MagMeDIP Kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/auto-methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020011</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1888" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1888" id="CartAdd/1888Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1888" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto MethylCap kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1888" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto MethylCap kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1887" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1887" id="CartAdd/1887Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1887" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MethylCap kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MethylCap kit',
'C02020010',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MethylCap kit',
'C02020010',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1887" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MethylCap kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/premium-bisulfite-kit-50-rxns"><img src="/img/grey-logo.jpg" alt="default alt" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02030030</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1892" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1892" id="CartAdd/1892Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1892" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Premium Bisulfite kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Premium Bisulfite kit',
'C02030030',
'240',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Premium Bisulfite kit',
'C02030030',
'240',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="premium-bisulfite-kit-50-rxns" data-reveal-id="cartModal-1892" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Premium Bisulfite kit</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1892',
'antibody_id' => null,
'name' => 'Premium Bisulfite kit',
'description' => '<p style="text-align: center;"><a href="https://www.diagenode.com/files/products/kits/Premium_Bisulfite_kit_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p style="text-align: center;"><strong>Make your Bisulfite conversion now in only 60 minutes !</strong></p>
<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C02030030',
'old_catalog_number' => '',
'sf_code' => 'C02030030-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'premium-bisulfite-kit-50-rxns',
'meta_title' => 'Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Premium Bisulfite kit',
'modified' => '2023-04-20 16:13:50',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '271',
'product_id' => '1979',
'related_id' => '1892'
),
'Image' => array()
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ' <span style="color:#CCC">(MAb-081-500)</span>'
$country_code = 'US'
$other_format = array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
'id' => '1914',
'product_id' => '1979',
'application_id' => '29'
)
)
$slugs = array(
(int) 0 => 'immunofluorescence'
)
$applications = array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'locale' => 'eng'
)
$description = '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>'
$name = 'IF'
$document = array(
'id' => '1118',
'name' => 'Datasheet 5-mC33D3 C15200081-10',
'description' => '<p><span>The 5-methylcytosine antibody (clone 33D3) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span></p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_5-mC33D3_C15200081-10.pdf',
'slug' => 'datasheet-5-mc33d3-c15200081-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-12-17 10:55:09',
'created' => '2020-12-17 10:55:09',
'ProductsDocument' => array(
'id' => '3063',
'product_id' => '1979',
'document_id' => '1118'
)
)
$sds = array(
'id' => '117',
'name' => '5-mC Antibody - clone 33D3 SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-03-13 15:43:48',
'created' => '2020-03-13 15:43:48',
'ProductsSafetySheet' => array(
'id' => '231',
'product_id' => '1979',
'safety_sheet_id' => '117'
)
)
$publication = array(
'id' => '976',
'name' => 'Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells.',
'authors' => 'Borgel J, Guibert S, Weber M.',
'description' => 'Methylated DNA immunoprecipitation (MeDIP) is an immunocapturing approach for unbiased enrichment of DNA that is methylated on cytosines. The principle is that genomic DNA is randomly sheared by sonication and immunoprecipitated with an antibody that specifically recognizes 5-methylcytidine (5mC), which can be combined with PCR or high-throughput analysis (microarrays, deep sequencing). The MeDIP technique has been originally used to generate DNA methylation profiles on a genome scale in mammals and plants. Here we provide an optimized version of the MeDIP protocol suitable for low amounts of DNA, which can be used to study DNA methylation in cellular populations available in small quantities.',
'date' => '0000-00-00',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22907495',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
'id' => '711',
'product_id' => '1979',
'publication_id' => '976'
)
)
$externalLink = ' <a href="http://www.ncbi.nlm.nih.gov/pubmed/22907495" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: campaign_id [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'product' => array(
'Product' => array(
'id' => '1979',
'antibody_id' => '631',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (sample size)',
'description' => '<p>Monoclonal antibody raised in mouse against 5-mC (<strong>5-methylcytosine</strong>) conjugated to ovalbumine (<strong>33D3 clone</strong>).</p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15200081-10',
'old_catalog_number' => 'MAb-081-010',
'sf_code' => 'C15200081-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10-ug-5-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:58:36',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '631',
'name' => '5-mC monoclonal antibody 33D3 (sample size)',
'description' => '',
'clonality' => '',
'isotype' => 'IgG1',
'lot' => 'RD-006',
'concentration' => '1.1 µg/µl',
'reactivity' => 'Human, mouse, rat, cow, other (wide range): positive',
'type' => 'Monoclonal. <br /><strong>MeDIP grade.</strong><br /><strong>MeDIP-seq grade</strong>.',
'purity' => 'Protein A purified monoclonal antibody.',
'classification' => '',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>MeDIP/MeDIP-seq <sup>*</sup></td>
<td>0.1 - 2 µg/IP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting **</td>
<td>1:300</td>
<td>Fig 3</td>
</tr>
<tr>
<td>IF</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
</tbody>
</table>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.1-5 µg per IP.</small></p>
<p><small><sup>**</sup> Dot blot was only performed to demonstrate the specificity. This antibody is not recommended for dot blot on biological samples.</small></p>
<p></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2023-05-11 15:21:00',
'created' => '2020-12-17 10:31:21',
'select_label' => '631 - 5-mC monoclonal antibody 33D3 (sample size) (RD-006 - 1.1 µg/µl - Human, mouse, rat, cow, other (wide range): positive - Protein A purified monoclonal antibody. - Mouse)'
),
'Slave' => array(),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
),
(int) 25 => array(
[maximum depth reached]
),
(int) 26 => array(
[maximum depth reached]
),
(int) 27 => array(
[maximum depth reached]
),
(int) 28 => array(
[maximum depth reached]
),
(int) 29 => array(
[maximum depth reached]
),
(int) 30 => array(
[maximum depth reached]
),
(int) 31 => array(
[maximum depth reached]
),
(int) 32 => array(
[maximum depth reached]
),
(int) 33 => array(
[maximum depth reached]
),
(int) 34 => array(
[maximum depth reached]
),
(int) 35 => array(
[maximum depth reached]
),
(int) 36 => array(
[maximum depth reached]
),
(int) 37 => array(
[maximum depth reached]
),
(int) 38 => array(
[maximum depth reached]
),
(int) 39 => array(
[maximum depth reached]
),
(int) 40 => array(
[maximum depth reached]
),
(int) 41 => array(
[maximum depth reached]
),
(int) 42 => array(
[maximum depth reached]
),
(int) 43 => array(
[maximum depth reached]
),
(int) 44 => array(
[maximum depth reached]
),
(int) 45 => array(
[maximum depth reached]
),
(int) 46 => array(
[maximum depth reached]
),
(int) 47 => array(
[maximum depth reached]
),
(int) 48 => array(
[maximum depth reached]
),
(int) 49 => array(
[maximum depth reached]
),
(int) 50 => array(
[maximum depth reached]
),
(int) 51 => array(
[maximum depth reached]
),
(int) 52 => array(
[maximum depth reached]
),
(int) 53 => array(
[maximum depth reached]
),
(int) 54 => array(
[maximum depth reached]
),
(int) 55 => array(
[maximum depth reached]
),
(int) 56 => array(
[maximum depth reached]
),
(int) 57 => array(
[maximum depth reached]
),
(int) 58 => array(
[maximum depth reached]
),
(int) 59 => array(
[maximum depth reached]
),
(int) 60 => array(
[maximum depth reached]
),
(int) 61 => array(
[maximum depth reached]
),
(int) 62 => array(
[maximum depth reached]
),
(int) 63 => array(
[maximum depth reached]
),
(int) 64 => array(
[maximum depth reached]
),
(int) 65 => array(
[maximum depth reached]
),
(int) 66 => array(
[maximum depth reached]
),
(int) 67 => array(
[maximum depth reached]
),
(int) 68 => array(
[maximum depth reached]
),
(int) 69 => array(
[maximum depth reached]
),
(int) 70 => array(
[maximum depth reached]
),
(int) 71 => array(
[maximum depth reached]
),
(int) 72 => array(
[maximum depth reached]
),
(int) 73 => array(
[maximum depth reached]
),
(int) 74 => array(
[maximum depth reached]
),
(int) 75 => array(
[maximum depth reached]
),
(int) 76 => array(
[maximum depth reached]
),
(int) 77 => array(
[maximum depth reached]
),
(int) 78 => array(
[maximum depth reached]
),
(int) 79 => array(
[maximum depth reached]
),
(int) 80 => array(
[maximum depth reached]
),
(int) 81 => array(
[maximum depth reached]
),
(int) 82 => array(
[maximum depth reached]
),
(int) 83 => array(
[maximum depth reached]
),
(int) 84 => array(
[maximum depth reached]
),
(int) 85 => array(
[maximum depth reached]
),
(int) 86 => array(
[maximum depth reached]
),
(int) 87 => array(
[maximum depth reached]
),
(int) 88 => array(
[maximum depth reached]
),
(int) 89 => array(
[maximum depth reached]
),
(int) 90 => array(
[maximum depth reached]
),
(int) 91 => array(
[maximum depth reached]
),
(int) 92 => array(
[maximum depth reached]
),
(int) 93 => array(
[maximum depth reached]
),
(int) 94 => array(
[maximum depth reached]
),
(int) 95 => array(
[maximum depth reached]
),
(int) 96 => array(
[maximum depth reached]
),
(int) 97 => array(
[maximum depth reached]
),
(int) 98 => array(
[maximum depth reached]
),
(int) 99 => array(
[maximum depth reached]
),
(int) 100 => array(
[maximum depth reached]
),
(int) 101 => array(
[maximum depth reached]
),
(int) 102 => array(
[maximum depth reached]
),
(int) 103 => array(
[maximum depth reached]
),
(int) 104 => array(
[maximum depth reached]
),
(int) 105 => array(
[maximum depth reached]
),
(int) 106 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul'
)
$language = 'en'
$meta_keywords = ''
$meta_description = '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.'
$meta_title = '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode'
$product = array(
'Product' => array(
'id' => '1979',
'antibody_id' => '631',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (sample size)',
'description' => '<p>Monoclonal antibody raised in mouse against 5-mC (<strong>5-methylcytosine</strong>) conjugated to ovalbumine (<strong>33D3 clone</strong>).</p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15200081-10',
'old_catalog_number' => 'MAb-081-010',
'sf_code' => 'C15200081-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10-ug-5-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-10) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP/MeDIP-seq, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:58:36',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '631',
'name' => '5-mC monoclonal antibody 33D3 (sample size)',
'description' => '',
'clonality' => '',
'isotype' => 'IgG1',
'lot' => 'RD-006',
'concentration' => '1.1 µg/µl',
'reactivity' => 'Human, mouse, rat, cow, other (wide range): positive',
'type' => 'Monoclonal. <br /><strong>MeDIP grade.</strong><br /><strong>MeDIP-seq grade</strong>.',
'purity' => 'Protein A purified monoclonal antibody.',
'classification' => '',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>MeDIP/MeDIP-seq <sup>*</sup></td>
<td>0.1 - 2 µg/IP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting **</td>
<td>1:300</td>
<td>Fig 3</td>
</tr>
<tr>
<td>IF</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
</tbody>
</table>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 0.1-5 µg per IP.</small></p>
<p><small><sup>**</sup> Dot blot was only performed to demonstrate the specificity. This antibody is not recommended for dot blot on biological samples.</small></p>
<p></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2023-05-11 15:21:00',
'created' => '2020-12-17 10:31:21',
'select_label' => '631 - 5-mC monoclonal antibody 33D3 (sample size) (RD-006 - 1.1 µg/µl - Human, mouse, rat, cow, other (wide range): positive - Protein A purified monoclonal antibody. - Mouse)'
),
'Slave' => array(),
'Group' => array(
'Group' => array(
'id' => '4',
'name' => 'C15200081',
'product_id' => '1980',
'modified' => '2016-02-17 17:32:42',
'created' => '2016-02-17 17:32:42'
),
'Master' => array(
'id' => '1980',
'antibody_id' => '630',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>Monoclonal antibody raised in mouse against </span><b>5-mC</b><span><span> </span>(</span><b>5-methylcytosine</b><span>) conjugated to ovalbumine (</span><b>33D3 clone</b><span>).</span></p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15200081-100',
'old_catalog_number' => 'MAb-081-100',
'sf_code' => 'C15200081-D001-000526',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '505',
'price_USD' => '575',
'price_GBP' => '450',
'price_JPY' => '79110',
'price_CNY' => '0',
'price_AUD' => '1438',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'October 27, 2020',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081) | Diagenode',
'meta_keywords' => '5-methylcytosine (5-mC),monoclonal antibody,Methylated DNA Immunoprecipitation',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2023-05-17 10:08:33',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1885',
'antibody_id' => null,
'name' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'description' => '<p><span>The Auro hMeDIP kit is designed for enrichment of hydroxymethylated DNA from fragmented genomic DNA samples for use in genome-wide methylation analysis. It features</span><span> a highly specific monoclonal antibody against </span><span>5-hydroxymethylcytosine (5-hmC) for the immunoprecipitation of hydroxymethylated DNA</span><span>. It includes control DNA and primers to assess the effiency of the assay. </span><span>Performing hydroxymethylation profiling with the hMeDIP kit is fast, reliable and highly specific.</span></p>',
'label1' => ' Characteristics',
'info1' => '<ul style="list-style-type: disc;">
<li><span>Robust enrichment & immunoprecipitation of hydroxymethylated DNA</span></li>
<li>Highly specific monoclonal antibody against 5-hmC<span> for reliable, reproducible results</span></li>
<li>Including control DNA and primers to <span>monitor the efficiency of the assay</span>
<ul style="list-style-type: circle;">
<li>5-hmC, 5-mC and unmethylated DNA sequences and primer pairs</li>
<li>Mouse primer pairs against Sfi1 targeting hydroxymethylated gene in mouse</li>
</ul>
</li>
</ul>
<ul style="list-style-type: disc;">
<li>Improved single-tube, magnetic bead-based protocol</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C02010034',
'old_catalog_number' => '',
'sf_code' => 'C02010034-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '630',
'price_USD' => '690',
'price_GBP' => '580',
'price_JPY' => '98690',
'price_CNY' => '',
'price_AUD' => '1725',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns',
'meta_title' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'meta_keywords' => '',
'meta_description' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'modified' => '2021-01-18 10:37:19',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1880',
'antibody_id' => null,
'name' => 'MagMeDIP Kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/magmedip-kit-manual-C02010020-21.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p> </p>
<div class="small-12 medium-4 large-4 columns"><center></center><center></center><center></center><center><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank"><img src="https://www.diagenode.com/img/banners/banner-nature-publication-580.png" alt="Click here to read more about MeDIP " caption="false" width="80%" /></a></center></div>
<div class="small-12 medium-8 large-8 columns">
<h3 style="text-align: justify;">Sensitive tumour detection and classification using plasma cell-free DNA methylomes<br /><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank">Read the publication</a></h3>
<h3 class="c-article-title u-h1" data-test="article-title" itemprop="name headline" style="text-align: justify;">Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA<br /><a href="https://www.nature.com/articles/s41596-019-0202-2" target="_blank" title="cfMeDIP-seq Nature Method">Read the method</a></h3>
</div>
<p></p>
<p></p>
<p></p>
<div class="row">
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
<div class="small-12 medium-8 large-8 columns"><br />
<p>Perform <strong>MeDIP</strong> (<strong>Me</strong>thylated <strong>D</strong>NA <strong>I</strong>mmuno<strong>p</strong>recipitation) followed by qPCR or NGS to estimate DNA methylation status of your sample using a highly sensitive 5-methylcytosine antibody. Our MagMeDIP kit contains high quality reagents to get the highest enrichment of methylated DNA with an optimized user-friendly protocol.</p>
</div>
</div>
<h3><span>Features</span></h3>
<ul>
<li>Starting DNA amount: <strong>10 ng – 1 µg</strong></li>
<li>Content: <strong>all reagents included</strong> for DNA extraction, immunoprecipitation (including the 5-mC antibody, spike-in controls and their corresponding qPCR primer pairs) as well as DNA isolation after IP.</li>
<li>Application: <strong>qPCR</strong> and <strong>NGS</strong></li>
<li>Robust method, <strong>superior enrichment</strong>, and easy-to-use protocol</li>
<li><strong>High reproducibility</strong> between replicates and repetitive experiments</li>
<li>Compatible with <strong>all species </strong></li>
</ul>',
'label1' => 'MagMeDIP workflow',
'info1' => '<p>DNA methylation occurs primarily as 5-methylcytosine (5-mC), and the Diagenode MagMeDIP Kit takes advantage of a specific antibody targeting this 5-mC to immunoprecipitate methylated DNA, which can be thereafter directly analyzed by qPCR or Next-Generation Sequencing (NGS).</p>
<h3><span>How it works</span></h3>
<p>In brief, after the cell collection and lysis, the genomic DNA is extracted, sheared, and then denatured. In the next step the antibody directed against 5 methylcytosine and antibody binding beads are used for immunoselection and immunoprecipitation of methylated DNA fragments. Then, the IP’d methylated DNA is isolated and can be used for any subsequent analysis as qPCR, amplification, hybridization on microarrays or next generation sequencing.</p>
<center><img src="https://www.diagenode.com/img/product/kits/MagMeDIP-workflow.png" width="70%" alt="5-methylcytosine" caption="false" /></center>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>',
'label2' => 'MeDIP-qPCR',
'info2' => '<p>The kit MagMeDIP contains all reagents necessary for a complete MeDIP-qPCR workflow. Two MagMeDIP protocols have been validated: for manual processing as well as for automated processing, using the Diagenode’s IP-Star Compact Automated System (please refer to the kit manual).</p>
<ul>
<li><strong>Complete kit</strong> including DNA extraction module, IP antibody and reagents, DNA isolation buffer</li>
<li><strong>Quality control of the IP:</strong> due to methylated and unmethylated DNA spike-in controls and their associated qPCR primers</li>
<li><strong>Easy to use</strong> with user-friendly magnetic beads and rack</li>
<li><strong>Highly validated protocol</strong></li>
<li>Automated protocol supplied</li>
</ul>
<center><img src="https://www.diagenode.com/img/product/kits/fig1-magmedipkit.png" width="85%" alt="Methylated DNA Immunoprecipitation" caption="false" /></center>
<p style="font-size: 0.9em;"><em><strong>Figure 1.</strong> Immunoprecipitation results obtained with Diagenode MagMeDIP Kit</em></p>
<p style="font-size: 0.9em;">MeDIP assays were performed manually using 1 µg or 50 ng gDNA from blood cells with the MagMeDIP kit (Diagenode). The IP was performed with the Methylated and Unmethylated spike-in controls included in the kit, together with the human DNA samples. The DNA was isolated/purified using DIB. Afterwards, qPCR was performed using the primer pairs included in this kit.</p>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>',
'label3' => 'MeDIP-seq',
'info3' => '<p>For DNA methylation analysis on the whole genome, MagMeDIP kit can be coupled with Next-Generation Sequencing. To perform MeDIP-sequencing we recommend the following strategy:</p>
<ul style="list-style-type: circle;">
<li>Choose a library preparation solution which is compatible with the starting amount of DNA you are planning to use (from 10 ng to 1 μg). It can be a home-made solution or a commercial one.</li>
<li>Choose the indexing system that fits your needs considering the following features:</li>
<ul>
<ul>
<ul>
<li>Single-indexing, combinatorial dual-indexing or unique dual-indexing</li>
<li>Number of barcodes</li>
<li>Full-length adaptors containing the barcodes or barcoding at the final amplification step</li>
<li>Presence / absence of Unique Molecular Identifiers (for PCR duplicates removal)</li>
</ul>
</ul>
</ul>
<li>Standard library preparation protocols are compatible with double-stranded DNA only, therefore the first steps of the library preparation (end repair, A-tailing, adaptor ligation and clean-up) will have to be performed on sheared DNA, before the IP.</li>
</ul>
<p style="padding-left: 30px;"><strong>CAUTION:</strong> As the immunoprecipitation step occurs at the middle of the library preparation workflow, single-tube solutions for library preparation are usually not compatible with MeDIP-sequencing.</p>
<ul style="list-style-type: circle;">
<li>For DNA isolation after the IP, we recommend using the <a href="https://www.diagenode.com/en/p/ipure-kit-v2-x24" title="IPure kit v2">IPure kit v2</a> (available separately, Cat. No. C03010014) instead of DNA isolation Buffer.</li>
</ul>
<ul style="list-style-type: circle;">
<li>Perform library amplification after the DNA isolation following the standard protocol of the chosen library preparation solution.</li>
</ul>
<h3><span>MeDIP-seq workflow</span></h3>
<center><img src="https://www.diagenode.com/img/product/kits/MeDIP-seq-workflow.png" width="110%" alt="MagMeDIP qPCR Kit x10 workflow" caption="false" /></center>
<h3><span>Example of results</span></h3>
<center><img src="https://www.diagenode.com/img/product/kits/medip-specificity.png" alt="MagMeDIP qPCR Kit Result" caption="false" width="951" height="488" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 1. qPCR analysis of external spike-in DNA controls (methylated and unmethylated) after IP.</strong> Samples were prepared using 1μg – 100ng -10ng sheared human gDNA with the MagMeDIP kit (Diagenode) and a commercially available library prep kit. DNA isolation after IP has been performed with IPure kit V2 (Diagenode).</p>
<p></p>
<p></p>
<center><img src="https://www.diagenode.com/img/product/kits/medip-saturation-analysis.png" alt=" MagMeDIP kit " caption="false" width="951" height="461" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 2. Saturation analysis.</strong> Clean reads were aligned to the human genome (hg19) using Burrows-Wheeler aligner (BWA) algorithm after which duplicated and unmapped reads were removed resulting in a mapping efficiency >98% for all samples. Quality and validity check of the mapped MeDIP-seq data was performed using MEDIPS R package. Saturation plots show that all sets of reads have sufficient complexity and depth to saturate the coverage profile of the reference genome and that this is reproducible between replicates and repetitive experiments (data shown for 50 ng gDNA input: left panel = replicate a, right panel = replicate b).</p>
<p></p>
<p></p>
<center><img src="https://www.diagenode.com/img/product/kits/medip-libraries-prep.png" alt="MagMeDIP x10 " caption="false" width="951" height="708" /></center>
<p></p>
<p style="font-size: 0.9em;"><strong>Figure 3. Sequencing profiles of MeDIP-seq libraries prepared from different starting amounts of sheared gDNA on the positive and negative methylated control regions.</strong> MeDIP-seq libraries were prepared from decreasing starting amounts of gDNA (1 μg (green), 50 ng (red), and 10ng (blue)) originating from human blood with the MagMeDIP kit (Diagenode) and a commercially available library prep kit. DNA isolation after IP has been performed with IPure kit V2 (Diagenode). IP and corresponding INPUT samples were sequenced on Illumina NovaSeq SP with 2x50 PE reads. The reads were mapped to the human genome (hg19) with bwa and the alignments were loaded into IGV (the tracks use an identical scale). The top IGV figure shows the TSH2B (also known as H2BC1) gene (marked by blue boxes in the bottom track) and its surroundings. The TSH2B gene is coding for a histone variant that does not occur in blood cells, and it is known to be silenced by methylation. Accordingly, we see a high coverage in the vicinity of this gene. The bottom IGV figure shows the GADPH locus (marked by blue boxes in the bottom track) and its surroundings. The GADPH gene is a highly active transcription region and should not be methylated, resulting in no reads accumulation following MeDIP-seq experiment.</p>
<p></p>
<ul>
<ul>
<script async="" src="https://edge.fullstory.com/s/fs.js" crossorigin="anonymous"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
</ul>
</ul>',
'format' => '48 rxns (IP)',
'catalog_number' => 'C02010021',
'old_catalog_number' => 'mc-magme-048',
'sf_code' => 'C02010021-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '745',
'price_USD' => '750',
'price_GBP' => '680',
'price_JPY' => '116705',
'price_CNY' => '',
'price_AUD' => '1875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'magmedip-kit-x48-48-rxns',
'meta_title' => 'MagMeDIP Kit for efficient immunoprecipitation of methylated DNA | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Perform Methylated DNA Immunoprecipitation (MeDIP) to estimate DNA methylation status of your sample using highly specific 5-mC antibody. This kit allows the preparation of cfMeDIP-seq libraries.',
'modified' => '2023-07-06 14:15:10',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1887',
'antibody_id' => null,
'name' => 'MethylCap kit',
'description' => '<p>The MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. The procedure has been adapted to both manual process or <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a>. Libraries of captured methylated DNA can be prepared for next-generation sequencing (NGS) by combining MBD technology with the <a href="https://www.diagenode.com/en/p/microplex-lib-prep-kit-v3-48-rxns">MicroPlex Library Preparation Kit v3</a>.</p>',
'label1' => 'Characteristics',
'info1' => '<ul style="list-style-type: circle;">
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>On-day protocol</strong></li>
<li><strong>NGS compatibility</strong></li>
</ul>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong></strong></p>
<p><strong></strong><strong>F</strong><strong>igure 1.</strong> Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).</p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020010',
'old_catalog_number' => 'AF-100-0048',
'sf_code' => 'C02020010-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'methylcap-kit-x48-48-rxns',
'meta_title' => 'MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'MethylCap kit x48',
'modified' => '2021-02-19 17:55:38',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '1892',
'antibody_id' => null,
'name' => 'Premium Bisulfite kit',
'description' => '<p style="text-align: center;"><a href="https://www.diagenode.com/files/products/kits/Premium_Bisulfite_kit_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p style="text-align: center;"><strong>Make your Bisulfite conversion now in only 60 minutes !</strong></p>
<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C02030030',
'old_catalog_number' => '',
'sf_code' => 'C02030030-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'premium-bisulfite-kit-50-rxns',
'meta_title' => 'Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Premium Bisulfite kit',
'modified' => '2023-04-20 16:13:50',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array([maximum depth reached])
)
),
'Application' => array(
(int) 0 => array(
'id' => '28',
'position' => '10',
'parent_id' => '40',
'name' => 'DB',
'description' => '<p>Dot blotting</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'dot-blotting',
'meta_keywords' => 'Dot blotting,Monoclonal & Polyclonal antibody,',
'meta_description' => 'Diagenode offers Monoclonal & Polyclonal antibodies for Dot blotting applications',
'meta_title' => 'Dot blotting Antibodies - Monoclonal & Polyclonal antibody | Diagenode',
'modified' => '2016-01-13 14:40:49',
'created' => '2015-07-08 13:45:05',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '23',
'position' => '40',
'parent_id' => '4',
'name' => 'DNA modifications',
'description' => '<p><span style="font-weight: 400;">T</span><span style="font-weight: 400;">he pattern of <strong>DNA modifications</strong> is critical for genome stability and the control of gene expression in the cell. Methylation of 5-cytosine (5-mC), one of the best-studied epigenetic marks, is carried out by the <strong>DNA methyltransferases</strong> DNMT3A and B and DNMT1. DNMT3A and DNMT3B are responsible for </span><i><span style="font-weight: 400;">de novo</span></i><span style="font-weight: 400;"> DNA methylation, whereas DNMT1 maintains existing methylation. 5-mC undergoes active demethylation which is performed by the <strong>Ten-Eleven Translocation</strong> (TET) familly of DNA hydroxylases. The latter consists of 3 members TET1, 2 and 3. All 3 members catalyze the conversion of <strong>5-methylcytosine</strong> (5-mC) into <strong>5-hydroxymethylcytosine</strong> (5-hmC), and further into <strong>5-formylcytosine</strong> (5-fC) and <strong>5-carboxycytosine</strong> (5-caC). 5-fC and 5-caC can be converted to unmodified cytosine by <strong>Thymine DNA Glycosylase</strong> (TDG). It is not yet clear if 5-hmC, 5-fC and 5-caC have specific functions or are simply intermediates in the demethylation of 5-mC.</span></p>
<p><span style="font-weight: 400;">DNA methylation is generally considered as a repressive mark and is usually associated with gene silencing. It is essential that the balance between DNA methylation and demethylation is precisely maintained. Dysregulation of DNA methylation may lead to many different human diseases and is often observed in cancer cells.</span></p>
<p><span style="font-weight: 400;">Diagenode offers highly validated antibodies against different proteins involved in DNA modifications as well as against the modified bases allowing the study of all steps and intermediates in the DNA methylation/demethylation pathway:</span></p>
<p><img src="https://www.diagenode.com/img/categories/antibodies/dna-methylation.jpg" height="599" width="816" /></p>
<p><strong>Diagenode exclusively sources the original 5-methylcytosine monoclonal antibody (clone 33D3).</strong></p>
<p>Check out the list below to see all proposed antibodies for DNA modifications.</p>
<p>Diagenode’s highly validated antibodies:</p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'dna-methylation-antibody',
'cookies_tag_id' => null,
'meta_keywords' => 'Monoclonal Antibodies,Polyclonal antibodies,DNA methylation,Diagenode',
'meta_description' => 'Diagenode offers Monoclonal and Polyclonal antibodies for DNA Methylation. The pattern of DNA modifications is critical for genome stability and the control of gene expression in the cell. ',
'meta_title' => 'DNA modifications - Monoclonal and Polyclonal Antibodies for DNA Methylation | Diagenode',
'modified' => '2020-08-03 15:25:37',
'created' => '2014-07-31 03:51:53',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '102',
'position' => '1',
'parent_id' => '4',
'name' => 'Sample size antibodies',
'description' => '<h1><strong>Validated epigenetics antibodies</strong> – care for a sample?<br /> </h1>
<p>Diagenode has partnered with leading epigenetics experts and numerous epigenetics consortiums to bring to you a validated and comprehensive collection of epigenetic antibodies. As an expert in epigenetics, we are committed to offering highly-specific antibodies validated for ChIP/ChIP-seq and many other applications. All batch-specific validation data is available on our website.<br /><a href="../categories/antibodies">Read about our expertise in antibody production</a>.</p>
<ul>
<li><strong>Focused</strong> - Diagenode's selection of antibodies is exclusively dedicated for epigenetic research. <a title="See the full collection." href="../categories/all-antibodies">See the full collection.</a></li>
<li><strong>Strict quality standards</strong> with rigorous QC and validation</li>
<li><strong>Classified</strong> based on level of validation for flexibility of application</li>
</ul>
<p>Existing sample sizes are listed below. We will soon expand our collection. Are you looking for a sample size of another antibody? Just <a href="mailto:agnieszka.zelisko@diagenode.com?Subject=Sample%20Size%20Request" target="_top">Contact us</a>.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => true,
'is_antibody' => true,
'slug' => 'sample-size-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => '5-hmC monoclonal antibody,CRISPR/Cas9 polyclonal antibody ,H3K36me3 polyclonal antibody,diagenode',
'meta_description' => 'Diagenode offers sample volume on selected antibodies for researchers to test, validate and provide confidence and flexibility in choosing from our wide range of antibodies ',
'meta_title' => 'Sample-size Antibodies | Diagenode',
'modified' => '2019-07-03 10:57:05',
'created' => '2015-10-27 12:13:34',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '15',
'name' => 'Monoclonal Antibody hMeDIP Kit for DNA Hydroxymethylation Studies',
'description' => '<p><span>There is substantial interest and speculation in the role of the “sixth DNA base,” 5-hydroxymethylcytosine (5-hmC), although its precise function has not yet been elucidated. Since its discovery in neuronal Purkinje, granule and ES cells, studies of this new modified DNA base have been limited by the lack of high-quality, validated tools and technologies that discriminate hydroxy-methylation from methylation in regulating genome expression. Obtaining a specific assay for 5-hmC is particularly important since standard bisulfite sequencing cannot distinguish between these two types of methylation. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Monoclonal_Antibody_hMeDIP_Kit_for_DNA_Hydroxymethylation_Studies_Poster.pdf',
'slug' => 'monoclonal-antibody-hmedip-kit-for-dna-hydroxymethylation-studies-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-09-29 13:42:52',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1118',
'name' => 'Datasheet 5-mC33D3 C15200081-10',
'description' => '<p><span>The 5-methylcytosine antibody (clone 33D3) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span></p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_5-mC33D3_C15200081-10.pdf',
'slug' => 'datasheet-5-mc33d3-c15200081-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-12-17 10:55:09',
'created' => '2020-12-17 10:55:09',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '250',
'name' => 'product/antibodies/antibody.png',
'alt' => 'Mouse IgG',
'modified' => '2020-11-27 07:00:09',
'created' => '2015-07-17 10:12:18',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4843',
'name' => 'Differentiation block in acute myeloid leukemia regulated by intronicsequences of FTO',
'authors' => 'Camera F. et al.',
'description' => '<p>Iroquois transcription factor gene IRX3 is highly expressed in 20–30\% of acute myeloid leukemia (AML) and contributes to the pathognomonic differentiation block. Intron 8 FTO sequences ∼220kB downstream of IRX3 exhibit histone acetylation, DNA methylation, and contacts with the IRX3 promoter, which correlate with IRX3 expression. Deletion of these intronic elements confirms a role in positively regulating IRX3. RNAseq revealed long non-coding (lnc) transcripts arising from this locus. FTO-lncAML knockdown (KD) induced differentiation of AML cells, loss of clonogenic activity, and reduced FTO intron 8:IRX3 promoter contacts. While both FTO-lncAML KD and IRX3 KD induced differentiation, FTO-lncAML but not IRX3 KD led to HOXA downregulation suggesting transcript activity in trans. FTO-lncAMLhigh AML samples expressed higher levels of HOXA and lower levels of differentiation genes. Thus, a regulatory module in FTO intron 8 consisting of clustered enhancer elements and a long non-coding RNA is active in human AML, impeding myeloid differentiation.</p>',
'date' => '2023-08-01',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2589004223013962',
'doi' => '10.1016/j.isci.2023.107319',
'modified' => '2023-08-01 14:14:01',
'created' => '2023-08-01 15:59:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4796',
'name' => 'Nicotinamide N-methyltransferase sustains a core epigenetic programthat promotes metastatic colonization in breast cancer.',
'authors' => 'Couto J.P. et al.',
'description' => '<p><span>Metastatic colonization of distant organs accounts for over 90% of deaths related to solid cancers, yet the molecular determinants of metastasis remain poorly understood. Here, we unveil a mechanism of colonization in the aggressive basal-like subtype of breast cancer that is driven by the NAD</span><sup>+</sup><span><span> </span>metabolic enzyme nicotinamide N-methyltransferase (NNMT). We demonstrate that NNMT imprints a basal genetic program into cancer cells, enhancing their plasticity. In line, NNMT expression is associated with poor clinical outcomes in patients with breast cancer. Accordingly, ablation of NNMT dramatically suppresses metastasis formation in pre-clinical mouse models. Mechanistically, NNMT depletion results in a methyl overflow that increases histone H3K9 trimethylation (H3K9me3) and DNA methylation at the promoters of PR/SET Domain-5 (PRDM5) and extracellular matrix-related genes. PRDM5 emerged in this study as a pro-metastatic gene acting via induction of cancer-cell intrinsic transcription of collagens. Depletion of PRDM5 in tumor cells decreases COL1A1 deposition and impairs metastatic colonization of the lungs. These findings reveal a critical activity of the NNMT-PRDM5-COL1A1 axis for cancer cell plasticity and metastasis in basal-like breast cancer.</span></p>',
'date' => '2023-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37259596',
'doi' => '10.15252/embj.2022112559',
'modified' => '2023-06-15 08:35:19',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4787',
'name' => 'The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.',
'authors' => 'Garcia-Gomez E. et al.',
'description' => '<p>Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the disease (control group). In addition, eight of the PCOS patients underwent intervention with metformin (1500 mg/day) and a carbohydrate-controlled diet (type and quantity) for three months. Clinical and metabolic parameters were determined, and RT-qPCR and MeDIP-qPCR were used to evaluate gene expression and DNA methylation levels, respectively. Decreased expression levels of , , and genes and increased DNA methylation levels of the promoter were found in the endometrium of PCOS patients compared to controls. After metformin and nutritional intervention, some metabolic and clinical variables improved in PCOS patients. This intervention was associated with increased expression of , , and genes and reduced DNA methylation levels of the promoter in the endometrium of PCOS women. Our preliminary findings suggest that metformin and a carbohydrate-controlled diet improve endometrial function in PCOS patients, partly by modulating DNA methylation of the gene promoter and the expression of genes implicated in endometrial receptivity and insulin signaling.</p>',
'date' => '2023-04-01',
'pmid' => 'https://doi.org/10.3390%2Fijms24076857',
'doi' => '10.3390/ijms24076857',
'modified' => '2023-06-12 08:58:33',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4777',
'name' => 'Epigenetic modifier alpha-ketoglutarate modulates aberrant gene bodymethylation and hydroxymethylation marks in diabetic heart.',
'authors' => 'Dhat R. et al.',
'description' => '<p>BACKGROUND: Diabetic cardiomyopathy (DCM) is a leading cause of death in diabetic patients. Hyperglycemic myocardial microenvironment significantly alters chromatin architecture and the transcriptome, resulting in aberrant activation of signaling pathways in a diabetic heart. Epigenetic marks play vital roles in transcriptional reprogramming during the development of DCM. The current study is aimed to profile genome-wide DNA (hydroxy)methylation patterns in the hearts of control and streptozotocin (STZ)-induced diabetic rats and decipher the effect of modulation of DNA methylation by alpha-ketoglutarate (AKG), a TET enzyme cofactor, on the progression of DCM. METHODS: Diabetes was induced in male adult Wistar rats with an intraperitoneal injection of STZ. Diabetic and vehicle control animals were randomly divided into groups with/without AKG treatment. Cardiac function was monitored by performing cardiac catheterization. Global methylation (5mC) and hydroxymethylation (5hmC) patterns were mapped in the Left ventricular tissue of control and diabetic rats with the help of an enrichment-based (h)MEDIP-sequencing technique by using antibodies specific for 5mC and 5hmC. Sequencing data were validated by performing (h)MEDIP-qPCR analysis at the gene-specific level, and gene expression was analyzed by qPCR. The mRNA and protein expression of enzymes involved in the DNA methylation and demethylation cycle were analyzed by qPCR and western blotting. Global 5mC and 5hmC levels were also assessed in high glucose-treated DNMT3B knockdown H9c2 cells. RESULTS: We found the increased expression of DNMT3B, MBD2, and MeCP2 with a concomitant accumulation of 5mC and 5hmC, specifically in gene body regions of diabetic rat hearts compared to the control. Calcium signaling was the most significantly affected pathway by cytosine modifications in the diabetic heart. Additionally, hypermethylated gene body regions were associated with Rap1, apelin, and phosphatidyl inositol signaling, while metabolic pathways were most affected by hyperhydroxymethylation. AKG supplementation in diabetic rats reversed aberrant methylation patterns and restored cardiac function. Hyperglycemia also increased 5mC and 5hmC levels in H9c2 cells, which was normalized by DNMT3B knockdown or AKG supplementation. CONCLUSION: This study demonstrates that reverting hyperglycemic damage to cardiac tissue might be possible by erasing adverse epigenetic signatures by supplementing epigenetic modulators such as AKG along with an existing antidiabetic treatment regimen.</p>',
'date' => '2023-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37101286',
'doi' => '10.1186/s13072-023-00489-4',
'modified' => '2023-06-12 09:20:54',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '4780',
'name' => 'Integrated analysis from multicentre studies identities RNAmethylation- related lncRNA risk stratification systems for glioma',
'authors' => 'Huang Fanxuan and Wang Xinyu and Zhong Junzhe and Chen Hao and Song Dan and Xu Tianye and Tian Kaifu and Sun Penggang and Sun Nan and Ma Wenbin and Liu Yuxiang andYu Daohan and Meng Xiangqi and Jiang Chuanlu and Xuan Hanwen and Qian Da an',
'description' => '<p>Gastric cancer (GC) is the fourth leading cause of cancer death worldwide. Due to the lack of effective chemotherapy methods for advanced gastric cancer and poor prognosis, the emergence of immunotherapy has brought new hope to gastric cancer. Further research is needed to improve the response rate to immunotherapy and identify the populations with potential benefits of immunotherapy. It is unclear whether m7G-related lncRNAs influence tumour immunity and the prognosis of immunotherapy.</p>',
'date' => '2023-03-02',
'pmid' => 'https://doi.org/10.21203%2Frs.3.rs-2815231%2Fv1',
'doi' => '10.21203/rs.3.rs-2815231/v1',
'modified' => '2023-06-13 09:25:12',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '4773',
'name' => 'The RNA m5C Methylase NSUN2 Modulates Corneal EpithelialWound Healing.',
'authors' => 'Luo G. et al.',
'description' => '<p>PURPOSE: The emerging epitranscriptomics offers insights into the physiopathological roles of various RNA modifications. The RNA methylase NOP2/Sun domain family member 2 (NSUN2) catalyzes 5-methylcytosine (m5C) modification of mRNAs. However, the role of NSUN2 in corneal epithelial wound healing (CEWH) remains unknown. Here we describe the functional mechanisms of NSUN2 in mediating CEWH. METHODS: RT-qPCR, Western blot, dot blot, and ELISA were used to determine the NSUN2 expression and overall RNA m5C level during CEWH. NSUN2 silencing or overexpression was performed to explore its involvement in CEWH both in vivo and in vitro. Multi-omics was integrated to reveal the downstream target of NSUN2. MeRIP-qPCR, RIP-qPCR, and luciferase assay, as well as in vivo and in vitro functional assays, clarified the molecular mechanism of NSUN2 in CEWH. RESULTS: The NSUN2 expression and RNA m5C level increased significantly during CEWH. NSUN2 knockdown significantly delayed CEWH in vivo and inhibited human corneal epithelial cells (HCEC) proliferation and migration in vitro, whereas NSUN2 overexpression prominently enhanced HCEC proliferation and migration. Mechanistically, we found that NSUN2 increased ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) translation through the binding of RNA m5C reader Aly/REF export factor. Accordingly, UHRF1 knockdown significantly delayed CEWH in vivo and inhibited HCEC proliferation and migration in vitro. Furthermore, UHRF1 overexpression effectively rescued the inhibitory effect of NSUN2 silencing on HCEC proliferation and migration. CONCLUSIONS: NSUN2-mediated m5C modification of UHRF1 mRNA modulates CEWH. This finding highlights the critical importance of this novel epitranscriptomic mechanism in control of CEWH.</p>',
'date' => '2023-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36862118',
'doi' => '10.1167/iovs.64.3.5',
'modified' => '2023-04-17 09:48:55',
'created' => '2023-04-14 13:41:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '4674',
'name' => 'Methylation and expression of glucocorticoid receptor exon-1 variants andFKBP5 in teenage suicide-completers.',
'authors' => 'Rizavi H. et al.',
'description' => '<p>A dysregulated hypothalamic-pituitary-adrenal (HPA) axis has repeatedly been demonstrated to play a fundamental role in psychiatric disorders and suicide, yet the mechanisms underlying this dysregulation are not clear. Decreased expression of the glucocorticoid receptor (GR) gene, which is also susceptible to epigenetic modulation, is a strong indicator of impaired HPA axis control. In the context of teenage suicide-completers, we have systematically analyzed the 5'UTR of the GR gene to determine the expression levels of all GR exon-1 transcript variants and their epigenetic state. We also measured the expression and the epigenetic state of the FK506-binding protein 51 (FKBP5/FKBP51), an important modulator of GR activity. Furthermore, steady-state DNA methylation levels depend upon the interplay between enzymes that promote DNA methylation and demethylation activities, thus we analyzed DNA methyltransferases (DNMTs), ten-eleven translocation enzymes (TETs), and growth arrest- and DNA-damage-inducible proteins (GADD45). Focusing on both the prefrontal cortex (PFC) and hippocampus, our results show decreased expression in specific GR exon-1 variants and a strong correlation of DNA methylation changes with gene expression in the PFC. FKBP5 expression is also increased in both areas suggesting a decreased GR sensitivity to cortisol binding. We also identified aberrant expression of DNA methylating and demethylating enzymes in both brain regions. These findings enhance our understanding of the complex transcriptional regulation of GR, providing evidence of epigenetically mediated reprogramming of the GR gene, which could lead to possible epigenetic influences that result in lasting modifications underlying an individual's overall HPA axis response and resilience to stress.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36781843',
'doi' => '10.1038/s41398-023-02345-1',
'modified' => '2023-04-14 09:26:37',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '4675',
'name' => 'Bridging biological cfDNA features and machine learning approaches.',
'authors' => 'Moser T. et al.',
'description' => '<p>Liquid biopsies (LBs), particularly using circulating tumor DNA (ctDNA), are expected to revolutionize precision oncology and blood-based cancer screening. Recent technological improvements, in combination with the ever-growing understanding of cell-free DNA (cfDNA) biology, are enabling the detection of tumor-specific changes with extremely high resolution and new analysis concepts beyond genetic alterations, including methylomics, fragmentomics, and nucleosomics. The interrogation of a large number of markers and the high complexity of data render traditional correlation methods insufficient. In this regard, machine learning (ML) algorithms are increasingly being used to decipher disease- and tissue-specific signals from cfDNA. Here, we review recent insights into biological ctDNA features and how these are incorporated into sophisticated ML applications.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36792446',
'doi' => '10.1016/j.tig.2023.01.004',
'modified' => '2023-04-14 09:28:00',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '4823',
'name' => 'Gene body DNA hydroxymethylation restricts the magnitude oftranscriptional changes during aging.',
'authors' => 'Occean J. R. et al.',
'description' => '<p>DNA hydroxymethylation (5hmC) is the most abundant oxidative derivative of DNA methylation (5mC) and is typically enriched at enhancers and gene bodies of transcriptionally active and tissue-specific genes. Although aberrant genomic 5hmC has been implicated in many age-related diseases, the functional role of the modification in aging remains largely unknown. Here, we report that 5hmC is stably enriched in multiple aged organs. Using the liver and cerebellum as model organs, we show that 5hmC accumulates in gene bodies associated with tissue-specific function and thereby restricts the magnitude of gene expression changes during aging. Mechanistically, we found that 5hmC decreases binding affinity of splicing factors compared to unmodified cytosine and 5mC, and is correlated with age-related alternative splicing events, suggesting RNA splicing as a potential mediator of 5hmC’s transcriptionally restrictive function. Furthermore, we show that various age-related contexts, such as prolonged quiescence and senescence, are partially responsible for driving the accumulation of 5hmC with age. We provide evidence that this age-related function is conserved in mouse and human tissues, and further show that the modification is altered by regimens known to modulate lifespan. Our findings reveal that 5hmC is a regulator of tissue-specific function and may play a role in regulating longevity.</p>',
'date' => '2023-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36824863',
'doi' => '10.1101/2023.02.15.528714',
'modified' => '2023-06-14 08:39:26',
'created' => '2023-06-13 22:16:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '4631',
'name' => 'Consistent DNA Hypomethylations in Prostate Cancer.',
'authors' => 'Araúzo-Bravo M.J. et al.',
'description' => '<p>With approximately 1.4 million men annually diagnosed with prostate cancer (PCa) worldwide, PCa remains a dreaded threat to life and source of devastating morbidity. In recent decades, a significant decrease in age-specific PCa mortality has been achieved by increasing prostate-specific antigen (PSA) screening and improving treatments. Nevertheless, upcoming, augmented recommendations against PSA screening underline an escalating disproportion between the benefit and harm of current diagnosis/prognosis and application of radical treatment standards. Undoubtedly, new potent diagnostic and prognostic tools are urgently needed to alleviate this tensed situation. They should allow a more reliable early assessment of the upcoming threat, in order to enable applying timely adjusted and personalized therapy and monitoring. Here, we present a basic study on an epigenetic screening approach by Methylated DNA Immunoprecipitation (MeDIP). We identified genes associated with hypomethylated CpG islands in three PCa sample cohorts. By adjusting our computational biology analyses to focus on single CpG-enriched 60-nucleotide-long DNA probes, we revealed numerous consistently differential methylated DNA segments in PCa. They were associated among other genes with and . These can be used for early discrimination, and might contribute to a new epigenetic tumor classification system of PCa. Our analysis shows that we can dissect short, differential methylated CpG-rich DNA fragments and combinations of them that are consistently present in all tumors. We name them tumor cell-specific differential methylated CpG dinucleotide signatures (TUMS).</p>',
'date' => '2022-12-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36613831',
'doi' => '10.3390/ijms24010386',
'modified' => '2023-03-28 09:03:47',
'created' => '2023-02-21 09:59:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '4534',
'name' => 'RNA 5-Methylcytosine Modification Regulates VegetativeDevelopment Associated with H3K27 Trimethylation inArabidopsis.',
'authors' => 'Zhang D.et al.',
'description' => '<p>Methylating RNA post-transcriptionally is emerging as a significant mechanism of gene regulation in eukaryotes. The crosstalk between RNA methylation and histone modification is critical for chromatin state and gene expression in mammals. However, it is not well understood mechanistically in plants. Here, the authors report a genome-wide correlation between RNA 5-cytosine methylation (m C) and histone 3 lysine27 trimethylation (H3K27me3) in Arabidopsis. The plant-specific Polycomb group (PcG) protein EMBRYONIC FLOWER1 (EMF1) plays dual roles as activators or repressors. Transcriptome-wide RNA m C profiling revealed that m C peaks are mostly enriched in chromatin regions that lacked H3K27me3 in both wild type and emf1 mutants. EMF1 repressed the expression of m C methyltransferase tRNA specific methyltransferase 4B (TRM4B) through H3K4me3, independent of PcG-mediated H3K27me3 mechanism. The 5-Cytosine methylation on targets is increased in emf1 mutants, thereby decreased the mRNA transcripts of photosynthesis and chloroplast genes. In addition, impairing EMF1 activity reduced H3K27me3 levels of PcG targets, such as starch genes, which are de-repressed in emf1 mutants. Both EMF1-mediated promotion and repression of gene activities via m C and H3K27me3 are required for normal vegetative growth. Collectively, t study reveals a previously undescribed epigenetic mechanism of RNA m C modifications and histone modifications to regulate gene expression in eukaryotes.</p>',
'date' => '2022-11-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36382558',
'doi' => '10.1002/advs.202204885',
'modified' => '2022-11-24 08:57:31',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '4541',
'name' => 'Cardiac epigenetic changes in VEGF signaling genes associates with myocardial microvascular rarefaction in experimental chronic kidney disease.',
'authors' => 'Eirin Alfonso and Chade Alejandro R',
'description' => '<p>BACKGROUND: Chronic kidney disease (CKD) is common in patients with heart failure, and often results in left ventricular diastolic dysfunction (LVDD). However, the mechanisms responsible for cardiac damage in CKD-LVDD remain to be elucidated. Epigenetic alterations may impose long-lasting effects on cellular transcription and function, but their exact role in CKD-LVDD is unknown. We investigate whether changes in cardiac site-specific DNA methylation profiles might be implicated in cardiac abnormalities in CKD-LVDD. METHODS: CKD-LVDD and normal control pigs (n=6 each) were studied for 14 weeks. Renal and cardiac hemodynamics were quantified by multidetector CT and echocardiography. In randomly selected pigs (n=3/group), cardiac site-specific 5-methylcytosine (5mC) immunoprecipitation (MeDIP)- and mRNA-sequencing (seq) was performed, followed by integrated (MeDiP-seq/mRNA-seq analysis), and confirmatory ex vivo studies. RESULTS: MeDIP-seq analysis revealed 261 genes with higher (fold-change>1.4; p<0.05) and 162 genes with lower (fold-change<0.7; p<0.05) 5mC levels in CKD-LVDD versus normal pigs, which were primarily implicated in vascular endothelial growth factor (VEGF)-related signaling and angiogenesis. Integrated MeDiP-seq/mRNA-seq analysis identified a select group of VEGF-related genes in which 5mC levels were higher, but mRNA expression lower in CKD-LVDD versus normal pigs. Cardiac VEGF signaling gene and VEGF protein expression was blunted in CKD-LVDD compared to controls and associated with decreased subendocardial microvascular density. CONCLUSIONS: Cardiac epigenetic changes in VEGF-related genes are associated with impaired angiogenesis and cardiac microvascular rarefaction in swine CKD-LVDD. These observations may assist in developing novel therapies to ameliorate cardiac damage in CKD-LVDD.</p>',
'date' => '2022-11-01',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/36367693/',
'doi' => '10.1152/ajpheart.00522.2022',
'modified' => '2022-11-25 09:03:31',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '4511',
'name' => 'The Arabidopsis APOLO and human UPAT sequence-unrelated longnoncoding RNAs can modulate DNA and histone methylation machineries inplants.',
'authors' => 'Fonouni-Farde C. et al.',
'description' => '<p>BACKGROUND: RNA-DNA hybrid (R-loop)-associated long noncoding RNAs (lncRNAs), including the Arabidopsis lncRNA AUXIN-REGULATED PROMOTER LOOP (APOLO), are emerging as important regulators of three-dimensional chromatin conformation and gene transcriptional activity. RESULTS: Here, we show that in addition to the PRC1-component LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), APOLO interacts with the methylcytosine-binding protein VARIANT IN METHYLATION 1 (VIM1), a conserved homolog of the mammalian DNA methylation regulator UBIQUITIN-LIKE CONTAINING PHD AND RING FINGER DOMAINS 1 (UHRF1). The APOLO-VIM1-LHP1 complex directly regulates the transcription of the auxin biosynthesis gene YUCCA2 by dynamically determining DNA methylation and H3K27me3 deposition over its promoter during the plant thermomorphogenic response. Strikingly, we demonstrate that the lncRNA UHRF1 Protein Associated Transcript (UPAT), a direct interactor of UHRF1 in humans, can be recognized by VIM1 and LHP1 in plant cells, despite the lack of sequence homology between UPAT and APOLO. In addition, we show that increased levels of APOLO or UPAT hamper VIM1 and LHP1 binding to YUCCA2 promoter and globally alter the Arabidopsis transcriptome in a similar manner. CONCLUSIONS: Collectively, our results uncover a new mechanism in which a plant lncRNA coordinates Polycomb action and DNA methylation through the interaction with VIM1, and indicates that evolutionary unrelated lncRNAs with potentially conserved structures may exert similar functions by interacting with homolog partners.</p>',
'date' => '2022-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36038910',
'doi' => '10.1186/s13059-022-02750-7',
'modified' => '2022-11-21 10:43:16',
'created' => '2022-11-15 09:26:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '4438',
'name' => 'A genome-wide screen reveals new regulators of the 2-cell-like cell state',
'authors' => 'Defossez Pierre-Antoine et al.',
'description' => '<p>In mammals, only the zygote and blastomeres of the early embryo are fully totipotent. This totipotency is mirrored in vitro by mouse "2-cell-like cells" (2CLCs), which appear at low frequency in cultures of Embryonic Stem cells (ESCs). Because totipotency is incompletely understood, we carried out a genomewide CRISPR KO screen in mouse ESCs, searching for mutants that reactivate the expression of Dazl, a robust 2-cell-like marker. Using secondary screens, we identify four mutants that reactivate not just Dazl, but also a broader 2-cell-like signature: the E3 ubiquitin ligase adaptor SPOP, the Zinc Finger transcription factor ZBTB14, MCM3AP, a component of the RNA processing complex TREX-2, and the lysine demethylase KDM5C. Functional experiments show how these factors link to known players of the 2 celllike state. These results extend our knowledge of totipotency, a key phase of organismal life.</p>',
'date' => '2022-06-01',
'pmid' => 'https://doi.org/10.21203%2Frs.3.rs-1561018%2Fv1',
'doi' => '10.21203/rs.3.rs-1561018/v1',
'modified' => '2022-09-28 09:23:42',
'created' => '2022-09-08 16:32:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '4553',
'name' => 'NSUN2-mediated RNA mC modification modulates uveal melanoma cellproliferation and migration.',
'authors' => 'Luo Guangying et al.',
'description' => '<p>RNA 5-methylcytosine (mC) is a widespread post-transcriptional modification involved in diverse biological processes through controlling RNA metabolism. However, its roles in uveal melanoma (UM) remain unknown. Here, we describe the biological roles and regulatory mechanisms of RNA mC in UM. Initially, we identified significantly elevated global RNA mC levels in both UM cells and tissue specimens using ELISA assay and dot blot analysis. Meanwhile, NOP2/Sun RNA methyltransferase family member 2 (NSUN2) was upregulated in both types of these samples, whereas NSUN2 knockdown significantly decreased RNA mC level. Such declines inhibited UM cell migration and suppressed cell proliferation through cell cycle G1 arrest. Furthermore, bioinformatic analyses, mC-RIP-qPCR, and luciferase assay identified β-Catenin (CTNNB1) as a direct target of NSUN2-mediated mC modification in UM cells. Additionally, overexpression of miR-124a in UM cells diminished NSUN2 expression levels indicating that it is an upstream regulator of this response. Our study suggests that NSUN2-mediated RNA mC methylation provides a potential novel target to improve the therapeutic management of UM pathogenesis.</p>',
'date' => '2022-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/35757999',
'doi' => '10.1080/15592294.2022.2088047',
'modified' => '2022-11-24 10:14:24',
'created' => '2022-11-24 08:49:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '4340',
'name' => 'Global DNA methylation and cellular 5-methylcytosine and H4acetylated patterns in primary and secondary dormant seeds of Capsellabursa-pastoris (L.) Medik. (shepherd's purse).',
'authors' => 'Gomez-Cabellos Sara et al.',
'description' => '<p>Despite the importance of dormancy and dormancy cycling for plants' fitness and life cycle phenology, a comprehensive characterization of the global and cellular epigenetic patterns across space and time in different seed dormancy states is lacking. Using Capsella bursa-pastoris (L.) Medik. (shepherd's purse) seeds with primary and secondary dormancy, we investigated the dynamics of global genomic DNA methylation and explored the spatio-temporal distribution of 5-methylcytosine (5-mC) and histone H4 acetylated (H4Ac) epigenetic marks. Seeds were imbibed at 30 °C in a light regime to maintain primary dormancy, or in darkness to induce secondary dormancy. An ELISA-based method was used to quantify DNA methylation, in relation to total genomic cytosines. Immunolocalization of 5-mC and H4Ac within whole seeds (i.e., including testa) was assessed with reference to embryo anatomy. Global DNA methylation levels were highest in prolonged (14 days) imbibed primary dormant seeds, with more 5-mC marked nuclei present only in specific parts of the seed (e.g., SAM and cotyledons). In secondary dormant seeds, global methylation levels and 5-mC signal where higher at 3 and 7 days than 1 or 14 days. With respect to acetylation, seeds had fewer H4Ac marked nuclei (e.g., SAM) in deeper dormant states, for both types of dormancy. However, the RAM still showed signal after 14 days of imbibition under dormancy-inducing conditions, suggesting a central role for the radicle/RAM in the response to perceived ambient changes and the adjustment of the seed dormancy state. Thus, we show that seed dormancy involves extensive cellular remodeling of DNA methylation and H4 acetylation.</p>',
'date' => '2022-05-01',
'pmid' => 'https://doi.org/10.1007%2Fs00709-021-01678-2',
'doi' => '10.1007/s00709-021-01678-2',
'modified' => '2022-06-20 09:19:49',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '4404',
'name' => 'Stella regulates the Development of Female Germline Stem Cells byModulating Chromatin Structure and DNA Methylation.',
'authors' => 'Hou Changliang et al.',
'description' => '<p>Female germline stem cells (FGSCs) have the ability to self-renew and differentiate into oocytes. , encoded by a maternal effect gene, plays an important role in oogenesis and early embryonic development. However, its function in FGSCs remains unclear. In this study, we showed that CRISPR/Cas9-mediated knockout of promoted FGSC proliferation and reduced the level of genome-wide DNA methylation of FGSCs. Conversely, overexpression led to the opposite results, and enhanced FGSC differentiation. We also performed an integrative analysis of chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), high-throughput genome-wide chromosome conformation capture (Hi-C), and use of our published epigenetic data. Results indicated that the binding sites of STELLA and active histones H3K4me3 and H3K27ac were enriched near the TAD boundaries. Hi-C analysis showed that overexpression attenuated the interaction within TADs, and interestingly enhanced the TAD boundary strength in STELLA-associated regions. Taking these findings together, our study not only reveals the role of in regulating DNA methylation and chromatin structure, but also provides a better understanding of FGSC development.</p>',
'date' => '2022-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9066111/',
'doi' => '10.7150/ijbs.69240',
'modified' => '2022-08-11 14:54:29',
'created' => '2022-08-11 12:14:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '4327',
'name' => 'Highly recurrent epimutations in gastric cancer CpG islandmethylator phenotypes and inflammation',
'authors' => 'Padmanabhan N. et al.',
'description' => '<p>Background CIMP (CpG island methylator phenotype) is an epigenetic molecular subtype, observed in multiple malignancies and associated with the epigenetic silencing of tumor suppressors. Currently, for most cancers including gastric cancer (GC), mechanisms underlying CIMP remain poorly understood. We sought to discover molecular contributors to CIMP in GC, by performing global DNA methylation, gene expression, and proteomics profiling across 14 gastric cell lines, followed by similar integrative analysis in 50 GC cell lines and 467 primary GCs. Results We identify the cystathionine beta-synthase enzyme (CBS) as a highly recurrent target of epigenetic silencing in CIMP GC. Likewise, we show that CBS epimutations are significantly associated with CIMP in various other cancers, occurring even in premalignant gastroesophageal conditions and longitudinally linked to clinical persistence. Of note, CRISPR deletion of CBS in normal gastric epithelial cells induces widespread DNA methylation changes that overlap with primary GC CIMP patterns. Reflecting its metabolic role as a gatekeeper interlinking the methionine and homocysteine cycles, CBS loss in vitro also causes reductions in the anti-inflammatory gasotransmitter hydrogen sulfide (H2S), with concomitant increase in NF-κB activity. In a murine genetic model of CBS deficiency, preliminary data indicate upregulated immune-mediated transcriptional signatures in the stomach. Conclusions Our results implicate CBS as a bi-faceted modifier of aberrant DNA methylation and inflammation in GC and highlights H2S donors as a potential new therapy for CBS-silenced lesions. Supplementary Information The online version contains supplementary material available at 10.1186/s13059-021-02375-2.</p>',
'date' => '2021-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/34074348',
'doi' => '10.1186/s13059-021-02375-2',
'modified' => '2022-08-03 16:01:40',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '4336',
'name' => 'LINE-1 transcription in round spermatids is associated with accretion of5-carboxylcytosine in their open reading frames',
'authors' => 'Blythe M. et al. ',
'description' => '<p>Chromatin of male and female gametes undergoes a number of reprogramming events during the transition from germ cell to embryonic developmental programs. Although the rearrangement of DNA methylation patterns occurring in the zygote has been extensively characterized, little is known about the dynamics of DNA modifications during spermatid maturation. Here, we demonstrate that the dynamics of 5-carboxylcytosine (5caC) correlate with active transcription of LINE-1 retroelements during murine spermiogenesis. We show that the open reading frames of active and evolutionary young LINE-1s are 5caC-enriched in round spermatids and 5caC is eliminated from LINE-1s and spermiogenesis-specific genes during spermatid maturation, being simultaneously retained at promoters and introns of developmental genes. Our results reveal an association of 5caC with activity of LINE-1 retrotransposons suggesting a potential direct role for this DNA modification in fine regulation of their transcription.</p>',
'date' => '2021-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/34099857',
'doi' => '10.1038/s42003-021-02217-8',
'modified' => '2022-08-03 16:17:04',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '4150',
'name' => 'Sensitive and reproducible cell-free methylome quantification with synthetic spike-in controls',
'authors' => 'Wilson, S.L. et al.',
'description' => '<p>Background. Cell-free methylated DNA immunoprecipitation-sequencing (cfMeDIP-seq) identifies genomic regions with DNA methylation, using a protocol adapted to work with low-input DNA samples and with cell-free DNA (cfDNA). This method allows for DNA methylation profiling of circulating tumour DNA in cancer patients’ blood samples. Such epigenetic profiling of circulating tumour DNA provides information about in which tissues tumour DNA originates, a key requirement of any test for early cancer detection. In addition, DNA methylation signatures provide prognostic information and can detect relapse. For robust quantitative comparisons between samples, immunoprecipitation enrichment methods like cfMeDIP-seq require normalization against common reference controls.</p>',
'date' => '2021-04-01',
'pmid' => 'https://doi.org/10.1101%2F2021.02.12.430289',
'doi' => '10.1101/2021.02.12.430289',
'modified' => '2022-01-13 15:16:23',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '3985',
'name' => 'Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes.',
'authors' => 'Nuzzo PV, Berchuck JE, Korthauer K, Spisak S, Nassar AH, Abou Alaiwi S, Chakravarthy A, Shen SY, Bakouny Z, Boccardo F, Steinharter J, Bouchard G, Curran CR, Pan W, Baca SC, Seo JH, Lee GM, Michaelson MD, Chang SL, Waikar SS, Sonpavde G, Irizarry RA, Pome',
'description' => '<p>Improving early cancer detection has the potential to substantially reduce cancer-related mortality. Cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) is a highly sensitive assay capable of detecting early-stage tumors. We report accurate classification of patients across all stages of renal cell carcinoma (RCC) in plasma (area under the receiver operating characteristic (AUROC) curve of 0.99) and demonstrate the validity of this assay to identify patients with RCC using urine cell-free DNA (cfDNA; AUROC of 0.86).</p>',
'date' => '2020-06-22',
'pmid' => 'http://www.pubmed.gov/32572266',
'doi' => '10.1038/s41591-020-0933-1',
'modified' => '2020-09-01 15:13:49',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '3984',
'name' => 'Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes.',
'authors' => 'Nassiri F, Chakravarthy A, Feng S, Shen SY, Nejad R, Zuccato JA, Voisin MR, Patil V, Horbinski C, Aldape K, Zadeh G, De Carvalho DD',
'description' => '<p>Definitive diagnosis of intracranial tumors relies on tissue specimens obtained by invasive surgery. Noninvasive diagnostic approaches provide an opportunity to avoid surgery and mitigate unnecessary risk to patients. In the present study, we show that DNA-methylation profiles from plasma reveal highly specific signatures to detect and accurately discriminate common primary intracranial tumors that share cell-of-origin lineages and can be challenging to distinguish using standard-of-care imaging.</p>',
'date' => '2020-06-22',
'pmid' => 'http://www.pubmed.gov/32572265',
'doi' => '10.1038/s41591-020-0932-2',
'modified' => '2020-09-01 15:14:45',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '4030',
'name' => 'AXR1 affects DNA methylation independently of its role in regulatingmeiotic crossover localization.',
'authors' => 'Christophorou, N and She, W and Long, J and Hurel, A and Beaubiat, S andIdir, Y and Tagliaro-Jahns, M and Chambon, A and Solier, V and Vezon, D andGrelon, M and Feng, X and Bouché, N and Mézard, C',
'description' => '<p>Meiotic crossovers (COs) are important for reshuffling genetic information between homologous chromosomes and they are essential for their correct segregation. COs are unevenly distributed along chromosomes and the underlying mechanisms controlling CO localization are not well understood. We previously showed that meiotic COs are mis-localized in the absence of AXR1, an enzyme involved in the neddylation/rubylation protein modification pathway in Arabidopsis thaliana. Here, we report that in axr1-/-, male meiocytes show a strong defect in chromosome pairing whereas the formation of the telomere bouquet is not affected. COs are also redistributed towards subtelomeric chromosomal ends where they frequently form clusters, in contrast to large central regions depleted in recombination. The CO suppressed regions correlate with DNA hypermethylation of transposable elements (TEs) in the CHH context in axr1-/- meiocytes. Through examining somatic methylomes, we found axr1-/- affects DNA methylation in a plant, causing hypermethylation in all sequence contexts (CG, CHG and CHH) in TEs. Impairment of the main pathways involved in DNA methylation is epistatic over axr1-/- for DNA methylation in somatic cells but does not restore regular chromosome segregation during meiosis. Collectively, our findings reveal that the neddylation pathway not only regulates hormonal perception and CO distribution but is also, directly or indirectly, a major limiting pathway of TE DNA methylation in somatic cells.</p>',
'date' => '2020-06-01',
'pmid' => 'http://www.pubmed.gov/32598340',
'doi' => '10.1371/journal.',
'modified' => '2020-12-16 17:58:51',
'created' => '2020-10-12 14:54:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '3951',
'name' => 'In vitro capture and characterization of embryonic rosette-stage pluripotency between naive and primed states.',
'authors' => 'Neagu A, van Genderen E, Escudero I, Verwegen L, Kurek D, Lehmann J, Stel J, Dirks RAM, van Mierlo G, Maas A, Eleveld C, Ge Y, den Dekker AT, Brouwer RWW, van IJcken WFJ, Modic M, Drukker M, Jansen JH, Rivron NC, Baart EB, Marks H, Ten Berge D',
'description' => '<p>Following implantation, the naive pluripotent epiblast of the mouse blastocyst generates a rosette, undergoes lumenogenesis and forms the primed pluripotent egg cylinder, which is able to generate the embryonic tissues. How pluripotency progression and morphogenesis are linked and whether intermediate pluripotent states exist remain controversial. We identify here a rosette pluripotent state defined by the co-expression of naive factors with the transcription factor OTX2. Downregulation of blastocyst WNT signals drives the transition into rosette pluripotency by inducing OTX2. The rosette then activates MEK signals that induce lumenogenesis and drive progression to primed pluripotency. Consequently, combined WNT and MEK inhibition supports rosette-like stem cells, a self-renewing naive-primed intermediate. Rosette-like stem cells erase constitutive heterochromatin marks and display a primed chromatin landscape, with bivalently marked primed pluripotency genes. Nonetheless, WNT induces reversion to naive pluripotency. The rosette is therefore a reversible pluripotent intermediate whereby control over both pluripotency progression and morphogenesis pivots from WNT to MEK signals.</p>',
'date' => '2020-05-01',
'pmid' => 'http://www.pubmed.gov/32367046',
'doi' => '10.1038/s41556-020-0508-x',
'modified' => '2020-08-17 09:55:37',
'created' => '2020-08-10 12:12:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '3834',
'name' => 'Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.',
'authors' => 'Liu J, An Z, Luo J, Li J, Li F, Zhang Z',
'description' => '<p>MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of transcriptome-wide profile and potential function of m5C in splicing remains to be elucidated due to lack of isoform level m5C quantification tool. RESULTS: We developed a computational package to quantify Epitranscriptomal RNA m5C at the transcript isoform level (named Episo). Episo consists of three tools, mapper, quant and Bisulfitefq, for mapping, quantifying, and simulating RNA-BisSeq data, respectively. The high accuracy of Episo was validated using an improved m5C-specific methylated RNA immunoprecipitation (meRIP) protocol, as well as a set of in silico experiments. By applying Episo to public human and mouse RNA-BisSeq data, we found that the RNA m5C is not evenly distributed among the transcript isoforms, implying the m5C may subject to be regulated at isoform level. AVAILABILITY: Episo is released under the GNU GPLv3+ license. The resource code Episo is freely accessible from https://github.com/liujunfengtop/Episo (with Tophat/cufflink) and https://github.com/liujunfengtop/Episo/tree/master/Episo_Kallisto (with Kallisto). SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.</p>',
'date' => '2019-12-03',
'pmid' => 'http://www.pubmed.gov/31794005',
'doi' => '10.1093/bioinformatics/btz900/5651015',
'modified' => '2020-02-25 13:26:22',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 25 => array(
'id' => '3827',
'name' => 'Intra- and inter-generational changes in the cortical DNA methylome in response to therapeutic intermittent hypoxia in mice.',
'authors' => 'Belmonte KCD, Harman JC, Lanson NA, Gidday JM',
'description' => '<p>Recent evidence from our lab documents functional resilience to retinal ischemic injury in untreated mice derived from parents exposed to repetitive hypoxic conditioning (RHC) prior to breeding. To begin to understand the epigenetic basis of this intergenerational protection, we used methylated DNA immunoprecipitation and sequencing to identify genes with differentially-methylated promoters (DMGPs) in the prefrontal cortex of mice treated directly with the same RHC stimulus (F0-RHC), and in the prefrontal cortex of their untreated F1-generation offspring (F1-*RHC). Subsequent bioinformatic analyses provided key mechanistic insights into how changes in gene expression secondary to promoter hypo- and hyper-methylation might afford such protection within and across generations. We found extensive changes in DNA methylation in both generations consistent with the expression of many survival-promoting genes, with twice the number of DMGPs in the cortex of F1*RHC mice relative to their F0 parents that were directly exposed to RHC. In contrast to our hypothesis that similar epigenetic modifications would be realized in the cortices of both F0-RHC and F1-*RHC mice, we instead found relatively few DMGPs common to both generations; in fact, each generation manifested expected injury resilience via distinctly unique gene expression profiles. Whereas in the cortex of F0-RHC mice, predicted protein-protein interactions reflected the activation of an anti-ischemic phenotype, networks activated in F1-*RHC cortex comprised networks indicative of a much broader cytoprotective phenotype. Altogether, our results suggest that the intergenerational transfer of an acquired phenotype to offspring does not necessarily require the faithful recapitulation of the conditioning-modified DNA methylome of the parent.</p>',
'date' => '2019-11-25',
'pmid' => 'http://www.pubmed.gov/31762411',
'doi' => '10.1152/physiolgenomics.00094.2019',
'modified' => '2020-02-25 13:35:09',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 26 => array(
'id' => '3814',
'name' => 'Lithium treatment reverses irradiation-induced changes in rodent neural progenitors and rescues cognition.',
'authors' => 'Zanni G, Goto S, Fragopoulou AF, Gaudenzi G, Naidoo V, Di Martino E, Levy G, Dominguez CA, Dethlefsen O, Cedazo-Minguez A, Merino-Serrais P, Stamatakis A, Hermanson O, Blomgren K',
'description' => '<p>Cranial radiotherapy in children has detrimental effects on cognition, mood, and social competence in young cancer survivors. Treatments harnessing hippocampal neurogenesis are currently of great relevance in this context. Lithium, a well-known mood stabilizer, has both neuroprotective, pro-neurogenic as well as antitumor effects, and in the current study we introduced lithium treatment 4 weeks after irradiation. Female mice received a single 4 Gy whole-brain radiation dose on postnatal day (PND) 21 and were randomized to 0.24% Li2CO chow or normal chow from PND 49 to 77. Hippocampal neurogenesis was assessed on PND 77, 91, and 105. We found that lithium treatment had a pro-proliferative effect on neural progenitors, but neuronal integration occurred only after it was discontinued. Also, the treatment ameliorated deficits in spatial learning and memory retention observed in irradiated mice. Gene expression profiling and DNA methylation analysis identified two novel factors related to the observed effects, Tppp, associated with microtubule stabilization, and GAD2/65, associated with neuronal signaling. Our results show that lithium treatment reverses irradiation-induced loss of hippocampal neurogenesis and cognitive impairment even when introduced long after the injury. We propose that lithium treatment should be intermittent in order to first make neural progenitors proliferate and then, upon discontinuation, allow them to differentiate. Our findings suggest that pharmacological treatment of cognitive so-called late effects in childhood cancer survivors is possible.</p>',
'date' => '2019-11-14',
'pmid' => 'http://www.pubmed.gov/31723242',
'doi' => '10.1038/s41380-019-0584-0',
'modified' => '2019-12-05 10:58:44',
'created' => '2019-12-02 15:25:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 27 => array(
'id' => '3773',
'name' => 'Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA.',
'authors' => 'Shen SY, Burgener JM, Bratman SV, De Carvalho DD',
'description' => '<p>Circulating cell-free DNA (cfDNA) comprises small DNA fragments derived from normal and tumor tissue that are released into the bloodstream. Recently, methylation profiling of cfDNA as a liquid biopsy tool has been gaining prominence due to the presence of tissue-specific markers in cfDNA. We have previously reported cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) as a sensitive, low-input, cost-efficient and bisulfite-free approach to profiling DNA methylomes of plasma cfDNA. cfMeDIP-seq is an extension of a previously published MeDIP-seq protocol and is adapted to allow for methylome profiling of samples with low input (ranging from 1 to 10 ng) of DNA, which is enabled by the addition of 'filler DNA' before immunoprecipitation. This protocol is not limited to plasma cfDNA; it can also be applied to other samples that are naturally sheared and at low availability (e.g., urinary cfDNA and cerebrospinal fluid cfDNA), and is potentially applicable to other applications beyond cancer detection, including prenatal diagnostics, cardiology and monitoring of immune response. The protocol presented here should enable any standard molecular laboratory to generate cfMeDIP-seq libraries from plasma cfDNA in ~3-4 d.</p>',
'date' => '2019-08-30',
'pmid' => 'http://www.pubmed.gov/31471598',
'doi' => '10.1038/s41596-019-0202-2',
'modified' => '2019-10-02 17:07:45',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 28 => array(
'id' => '3763',
'name' => 'Silencing of tumor-suppressive NR_023387 in renal cell carcinoma via promoter hypermethylation and HNF4A deficiency.',
'authors' => 'Zhou H, Guo L, Yao W, Shi R, Yu G, Xu H, Ye Z',
'description' => '<p>Dysregulation of the epigenetic status of long noncoding RNAs (lncRNAs) has been linked to diverse human diseases including human cancers. However, the landscape of the whole-genome methylation profile of lncRNAs and the precise roles of these lncRNAs remain elusive in renal cell carcinoma (RCC). We first examined lncRNA expression profiles in RCC tissues and corresponding adjacent normal tissues (NTs) to identify the lncRNA signature of RCC, then lncRNA Promoter Microarray was performed to depict the whole-genome methylation profile of lncRNAs in RCC. Combined analysis of the lncRNAs expression profiles and lncRNAs Promoter Microarray identified a series of downregulated lncRNAs with hypermethylated promoter regions, including NR_023387. Quantitative real-time polymerase chain reaction (RT-PCR) implied that NR_023387 was significantly downregulated in RCC tissues and cell lines, and lower expression of NR_023387 was correlated with shorter overall survival. Methylation-specific PCR, MassARRAY, and demethylation drug treatment indicated that hypermethylation in the NR_023387 promoter contributed to its silencing in RCC. Besides, HNF4A regulated the expression of NR_023387 via transcriptional activation. Functional experiments demonstrated NR_023387 exerted tumor-suppressive roles in RCC via suppressing the proliferation, migration, invasion, tumor growth, and metastasis of RCC. Furthermore, we identified MGP as a putative downstream molecule of NR_023387, which promoted the epithelial-mesenchymal transition of RCC cells. Our study provides the first whole-genome lncRNA methylation profile in RCC. Our combined analysis identifies a tumor-suppressive and prognosis-related lncRNA NR_023387, which is silenced in RCC via promoter hypermethylation and HNF4A deficiency, and may exert its tumor-suppressive roles by downregulating the oncogenic MGP.</p>',
'date' => '2019-08-20',
'pmid' => 'http://www.pubmed.gov/31432508',
'doi' => '10.1002/jcp.29115',
'modified' => '2019-10-03 10:02:27',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 29 => array(
'id' => '3770',
'name' => 'Epitranscriptomic Addition of mC to HIV-1 Transcripts Regulates Viral Gene Expression.',
'authors' => 'Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR',
'description' => '<p>How the covalent modification of mRNA ribonucleotides, termed epitranscriptomic modifications, alters mRNA function remains unclear. One issue has been the difficulty of quantifying these modifications. Using purified HIV-1 genomic RNA, we show that this RNA bears more epitranscriptomic modifications than the average cellular mRNA, with 5-methylcytosine (mC) and 2'O-methyl modifications being particularly prevalent. The methyltransferase NSUN2 serves as the primary writer for mC on HIV-1 RNAs. NSUN2 inactivation inhibits not only mC addition to HIV-1 transcripts but also viral replication. This inhibition results from reduced HIV-1 protein, but not mRNA, expression, which in turn correlates with reduced ribosome binding to viral mRNAs. In addition, loss of mC dysregulates the alternative splicing of viral RNAs. These data identify mC as a post-transcriptional regulator of both splicing and function of HIV-1 mRNA, thereby affecting directly viral gene expression.</p>',
'date' => '2019-08-14',
'pmid' => 'http://www.pubmed.gov/31415754',
'doi' => '10.1016/j.chom.2019.07.005',
'modified' => '2019-10-03 09:18:50',
'created' => '2019-10-02 16:16:55',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 30 => array(
'id' => '3741',
'name' => 'Aberrant expression of imprinted lncRNA MEG8 causes trophoblast dysfunction and abortion.',
'authors' => 'Sheng F, Sun N, Ji Y, Ma Y, Ding H, Zhang Q, Yang F, Li W',
'description' => '<p>Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs whose nucleotides are longer than 200 bp. Previous studies have shown that they play an important regulatory role in many developmental processes and biological pathways. However, the contributions of lncRNAs to placental development are largely unknown. Here, our study aimed to investigate the lncRNA expression signatures in placental development by performing a microarray lncRNA screen. Placental samples were obtained from pregnant C57BL/6 female mice at three key developmental time points (embryonic day E7.5, E13.5, and E19.5). Microarrays were used to analyze the differential expression of lncRNAs during placental development. In addition to the genomic imprinting region and the dynamic DNA methylation status during placental development, we screened imprinted lncRNAs whose expression was controlled by DNA methylation during placental development. We found that the imprinted lncRNA Rian may play an important role during placental development. Its homologous sequence lncRNA MEG8 (RIAN) was abnormally highly expressed in human spontaneous abortion villi. Upregulation of MEG8 expression in trophoblast cell lines decreased cell proliferation and invasion, whereas downregulation of MEG8 expression had the opposite effect. Furthermore, DNA methylation results showed that the methylation of the MEG8 promoter region was increased in spontaneous abortion villi. There was dynamic spatiotemporal expression of imprinted lncRNAs during placental development. The imprinted lncRNA MEG8 is involved in the regulation of early trophoblast cell function. Promoter methylation abnormalities can cause trophoblastic cell defects, which may be one of the factors that occurs in early unexplained spontaneous abortion.</p>',
'date' => '2019-07-02',
'pmid' => 'http://www.pubmed.gov/31265183',
'doi' => '10.1002/jcb.29002',
'modified' => '2019-08-06 16:45:53',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 31 => array(
'id' => '3731',
'name' => 'Defining UHRF1 Domains that Support Maintenance of Human Colon Cancer DNA Methylation and Oncogenic Properties.',
'authors' => 'Kong X, Chen J, Xie W, Brown SM, Cai Y, Wu K, Fan D, Nie Y, Yegnasubramanian S, Tiedemann RL, Tao Y, Chiu Yen RW, Topper MJ, Zahnow CA, Easwaran H, Rothbart SB, Xia L, Baylin SB',
'description' => '<p>UHRF1 facilitates the establishment and maintenance of DNA methylation patterns in mammalian cells. The establishment domains are defined, including E3 ligase function, but the maintenance domains are poorly characterized. Here, we demonstrate that UHRF1 histone- and hemimethylated DNA binding functions, but not E3 ligase activity, maintain cancer-specific DNA methylation in human colorectal cancer (CRC) cells. Disrupting either chromatin reader activity reverses DNA hypermethylation, reactivates epigenetically silenced tumor suppressor genes (TSGs), and reduces CRC oncogenic properties. Moreover, an inverse correlation between high UHRF1 and low TSG expression tracks with CRC progression and reduced patient survival. Defining critical UHRF1 domain functions and its relationship with CRC prognosis suggests directions for, and value of, targeting this protein to develop therapeutic DNA demethylating agents.</p>',
'date' => '2019-04-15',
'pmid' => 'http://www.pubmed.gov/30956060',
'doi' => '10.1016/j.ccell.2019.03.003',
'modified' => '2019-08-07 09:14:54',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 32 => array(
'id' => '3729',
'name' => 'Tricarboxylic Acid Cycle Activity and Remodeling of Glycerophosphocholine Lipids Support Cytokine Induction in Response to Fungal Patterns.',
'authors' => 'Márquez S, Fernández JJ, Mancebo C, Herrero-Sánchez C, Alonso S, Sandoval TA, Rodríguez Prados M, Cubillos-Ruiz JR, Montero O, Fernández N, Sánchez Crespo M',
'description' => '<p>Increased glycolysis parallels immune cell activation, but the role of pyruvate remains largely unexplored. We found that stimulation of dendritic cells with the fungal surrogate zymosan causes decreases of pyruvate, citrate, itaconate, and α-ketoglutarate, while increasing oxaloacetate, succinate, lactate, oxygen consumption, and pyruvate dehydrogenase activity. Expression of IL10 and IL23A (the gene encoding the p19 chain of IL-23) depended on pyruvate dehydrogenase activity. Mechanistically, pyruvate reinforced histone H3 acetylation, and acetate rescued the effect of mitochondrial pyruvate carrier inhibition, most likely because it is a substrate of the acetyl-CoA producing enzyme ACSS2. Mice lacking the receptor of the lipid mediator platelet-activating factor (PAF; 1-O-hexadecyl-2-acetyl-sn-glycero-3-phosphocholine) showed reduced production of IL-10 and IL-23 that is explained by the requirement of acetyl-CoA for PAF biosynthesis and its ensuing autocrine function. Acetyl-CoA therefore intertwines fatty acid remodeling of glycerophospholipids and energetic metabolism during cytokine induction.</p>',
'date' => '2019-04-09',
'pmid' => 'http://www.pubmed.gov/30970255',
'doi' => '10.1016/j.celrep.2019.03.033',
'modified' => '2019-08-07 09:15:46',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 33 => array(
'id' => '3693',
'name' => 'Increased Serine and One-Carbon Pathway Metabolism by PKCλ/ι Deficiency Promotes Neuroendocrine Prostate Cancer.',
'authors' => 'Reina-Campos M, Linares JF, Duran A, Cordes T, L'Hermitte A, Badur MG, Bhangoo MS, Thorson PK, Richards A, Rooslid T, Garcia-Olmo DC, Nam-Cha SY, Salinas-Sanchez AS, Eng K, Beltran H, Scott DA, Metallo CM, Moscat J, Diaz-Meco MT',
'description' => '<p>Increasingly effective therapies targeting the androgen receptor have paradoxically promoted the incidence of neuroendocrine prostate cancer (NEPC), the most lethal subtype of castration-resistant prostate cancer (PCa), for which there is no effective therapy. Here we report that protein kinase C (PKC)λ/ι is downregulated in de novo and during therapy-induced NEPC, which results in the upregulation of serine biosynthesis through an mTORC1/ATF4-driven pathway. This metabolic reprogramming supports cell proliferation and increases intracellular S-adenosyl methionine (SAM) levels to feed epigenetic changes that favor the development of NEPC characteristics. Altogether, we have uncovered a metabolic vulnerability triggered by PKCλ/ι deficiency in NEPC, which offers potentially actionable targets to prevent therapy resistance in PCa.</p>',
'date' => '2019-03-18',
'pmid' => 'http://www.pubmed.gov/30827887',
'doi' => '10.1016/j.ccell.2019.01.018',
'modified' => '2019-06-28 13:49:24',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 34 => array(
'id' => '3660',
'name' => 'Global distribution of DNA hydroxymethylation and DNA methylation in chronic lymphocytic leukemia.',
'authors' => 'Wernig-Zorc S, Yadav MP, Kopparapu PK, Bemark M, Kristjansdottir HL, Andersson PO, Kanduri C, Kanduri M',
'description' => '<p>BACKGROUND: Chronic lymphocytic leukemia (CLL) has been a good model system to understand the functional role of 5-methylcytosine (5-mC) in cancer progression. More recently, an oxidized form of 5-mC, 5-hydroxymethylcytosine (5-hmC) has gained lot of attention as a regulatory epigenetic modification with prognostic and diagnostic implications for several cancers. However, there is no global study exploring the role of 5-hydroxymethylcytosine (5-hmC) levels in CLL. Herein, using mass spectrometry and hMeDIP-sequencing, we analysed the dynamics of 5-hmC during B cell maturation and CLL pathogenesis. RESULTS: We show that naïve B-cells had higher levels of 5-hmC and 5-mC compared to non-class switched and class-switched memory B-cells. We found a significant decrease in global 5-mC levels in CLL patients (n = 15) compared to naïve and memory B cells, with no changes detected between the CLL prognostic groups. On the other hand, global 5-hmC levels of CLL patients were similar to memory B cells and reduced compared to naïve B cells. Interestingly, 5-hmC levels were increased at regulatory regions such as gene-body, CpG island shores and shelves and 5-hmC distribution over the gene-body positively correlated with degree of transcriptional activity. Importantly, CLL samples showed aberrant 5-hmC and 5-mC pattern over gene-body compared to well-defined patterns in normal B-cells. Integrated analysis of 5-hmC and RNA-sequencing from CLL datasets identified three novel oncogenic drivers that could have potential roles in CLL development and progression. CONCLUSIONS: Thus, our study suggests that the global loss of 5-hmC, accompanied by its significant increase at the gene regulatory regions, constitute a novel hallmark of CLL pathogenesis. Our combined analysis of 5-mC and 5-hmC sequencing provided insights into the potential role of 5-hmC in modulating gene expression changes during CLL pathogenesis.</p>',
'date' => '2019-01-07',
'pmid' => 'http://www.pubmed.gov/30616658',
'doi' => '10.1186/s13072‑018‑0252‑7',
'modified' => '2019-07-01 11:46:16',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 35 => array(
'id' => '3730',
'name' => 'Transcriptome-Wide Mapping 5-Methylcytosine by mC RNA Immunoprecipitation Followed by Deep Sequencing in Plant.',
'authors' => 'Gu X, Liang Z',
'description' => '<p>Transcriptome-wide mapping RNA modification is crucial to understand the distribution and function of RNA modifications. Here, we describe a protocol to transcriptome-wide mapping 5-methylcytosine (mC) in plant, by a RNA immunoprecipitation followed by deep sequencing (mC-RIP-seq) approach. The procedure includes RNA extraction, fragmentation, RNA immunoprecipitation, and library construction.</p>',
'date' => '2019-01-01',
'pmid' => 'http://www.pubmed.gov/30945199',
'doi' => '10.1007/978-1-4939-9045-0_24,',
'modified' => '2019-08-07 10:21:37',
'created' => '2019-07-31 13:35:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 36 => array(
'id' => '3584',
'name' => 'Sensitivity of pituitary gonadotropes to hyperglycemia leads to epigenetic aberrations and reduced follicle-stimulating hormone levels.',
'authors' => 'Feldman A, Saleh A, Pnueli L, Qiao S, Shlomi T, Boehm U, Melamed P',
'description' => '<p>The connection between metabolism and reproductive function is well recognized, and we hypothesized that the pituitary gonadotropes, which produce luteinizing hormone and follicle-stimulating hormone (FSH), mediate some of the effects directly via insulin-independent glucose transporters, which allow continued glucose metabolism during hyperglycemia. We found that glucose transporter 1 is the predominant glucose transporter in primary gonadotropes and a gonadotrope precursor-derived cell line, and both are responsive to culture in high glucose; moreover, metabolite levels were altered in the cell line. Several of the affected metabolites are cofactors for chromatin-modifying enzymes, and in the gonadotrope precursor-derived cell line, we recorded global changes in histone acetylation and methylation, decreased DNA methylation, and increased hydroxymethylation, some of which did not revert to basal levels after cells were returned to normal glucose. Despite this weakening of epigenetic-mediated repression seen in the model cell line, FSH β-subunit ( Fshb) mRNA levels in primary gonadotropes were significantly reduced, apparently due in part to increased autocrine/paracrine effects of inhibin. However, unlike thioredoxin interacting protein and inhibin subunit α, Fshb mRNA levels did not recover after the return of cells to normal glucose. The effect on Fshb expression was also seen in 2 hyperglycemic mouse models, and levels of circulating FSH, required for follicle growth and development, were reduced. Thus, hyperglycemia seems to target the pituitary gonadotropes directly, and the likely extensive epigenetic changes are sensed acutely by Fshb. This scenario would explain clinical findings in which, even after restoration of optimal blood glucose levels, fertility often remains adversely affected. However, the relative accessibility of the pituitary provides a possible target for treatment, particularly crucial in the young in which hyperglycemia is increasingly common and fertility most relevant.-Feldman, A., Saleh, A., Pnueli, L., Qiao, S., Shlomi, T., Boehm, U., Melamed, P. Sensitivity of pituitary gonadotropes to hyperglycemia leads to epigenetic aberrations and reduced follicle-stimulating hormone levels.</p>',
'date' => '2018-12-27',
'pmid' => 'http://www.pubmed.gov/30074825',
'doi' => '10.1096/fj.201800943R',
'modified' => '2019-04-17 15:48:51',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 37 => array(
'id' => '3430',
'name' => 'Sensitive tumour detection and classification using plasma cell-free DNA methylomes.',
'authors' => 'Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, Kis O, Zhao Z, Spreafico A, Medina TDS, Wang Y, Roulois D, Ettayebi I, Chen Z, Chow S, Murphy T, Arruda A, O'Kane GM, Liu J, Mansour M, McPher',
'description' => '<p>The use of liquid biopsies for cancer detection and management is rapidly gaining prominence. Current methods for the detection of circulating tumour DNA involve sequencing somatic mutations using cell-free DNA, but the sensitivity of these methods may be low among patients with early-stage cancer given the limited number of recurrent mutations. By contrast, large-scale epigenetic alterations-which are tissue- and cancer-type specific-are not similarly constrained and therefore potentially have greater ability to detect and classify cancers in patients with early-stage disease. Here we develop a sensitive, immunoprecipitation-based protocol to analyse the methylome of small quantities of circulating cell-free DNA, and demonstrate the ability to detect large-scale DNA methylation changes that are enriched for tumour-specific patterns. We also demonstrate robust performance in cancer detection and classification across an extensive collection of plasma samples from several tumour types. This work sets the stage to establish biomarkers for the minimally invasive detection, interception and classification of early-stage cancers based on plasma cell-free DNA methylation patterns.</p>',
'date' => '2018-11-14',
'pmid' => 'http://www.pubmed.gov/30429608',
'doi' => '10.1038/s41586-018-0703-0',
'modified' => '2019-06-11 16:22:54',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 38 => array(
'id' => '3421',
'name' => 'Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate.',
'authors' => 'Sardina JL, Collombet S, Tian TV, Gómez A, Di Stefano B, Berenguer C, Brumbaugh J, Stadhouders R, Segura-Morales C, Gut M, Gut IG, Heath S, Aranda S, Di Croce L, Hochedlinger K, Thieffry D, Graf T',
'description' => '<p>Here, we report DNA methylation and hydroxymethylation dynamics at nucleotide resolution using C/EBPα-enhanced reprogramming of B cells into induced pluripotent cells (iPSCs). We observed successive waves of hydroxymethylation at enhancers, concomitant with a decrease in DNA methylation, suggesting active demethylation. Consistent with this finding, ablation of the DNA demethylase Tet2 almost completely abolishes reprogramming. C/EBPα, Klf4, and Tfcp2l1 each interact with Tet2 and recruit the enzyme to specific DNA sites. During reprogramming, some of these sites maintain high levels of 5hmC, and enhancers and promoters of key pluripotency factors become demethylated as early as 1 day after Yamanaka factor induction. Surprisingly, methylation changes precede chromatin opening in distinct chromatin regions, including Klf4 bound sites, revealing a pioneer factor activity associated with alternation in DNA methylation. Rapid changes in hydroxymethylation similar to those in B cells were also observed during compound-accelerated reprogramming of fibroblasts into iPSCs, highlighting the generality of our observations.</p>',
'date' => '2018-11-01',
'pmid' => 'http://www.pubmed.gov/30220521',
'doi' => '10.1016/j.stem.2018.08.016',
'modified' => '2018-12-31 11:16:24',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 39 => array(
'id' => '3409',
'name' => 'Oxidative stress in sperm affects the epigenetic reprogramming in early embryonic development.',
'authors' => 'Wyck S, Herrera C, Requena CE, Bittner L, Hajkova P, Bollwein H, Santoro R',
'description' => '<p>BACKGROUND: Reactive oxygen species (ROS)-induced oxidative stress is well known to play a major role in male infertility. Sperm are sensitive to ROS damaging effects because as male germ cells form mature sperm they progressively lose the ability to repair DNA damage. However, how oxidative DNA lesions in sperm affect early embryonic development remains elusive. RESULTS: Using cattle as model, we show that fertilization using sperm exposed to oxidative stress caused a major developmental arrest at the time of embryonic genome activation. The levels of DNA damage response did not directly correlate with the degree of developmental defects. The early cellular response for DNA damage, γH2AX, is already present at high levels in zygotes that progress normally in development and did not significantly increase at the paternal genome containing oxidative DNA lesions. Moreover, XRCC1, a factor implicated in the last step of base excision repair (BER) pathway, was recruited to the damaged paternal genome, indicating that the maternal BER machinery can repair these DNA lesions induced in sperm. Remarkably, the paternal genome with oxidative DNA lesions showed an impairment of zygotic active DNA demethylation, a process that previous studies linked to BER. Quantitative immunofluorescence analysis and ultrasensitive LC-MS-based measurements revealed that oxidative DNA lesions in sperm impair active DNA demethylation at paternal pronuclei, without affecting 5-hydroxymethylcytosine (5hmC), a 5-methylcytosine modification that has been implicated in paternal active DNA demethylation in mouse zygotes. Thus, other 5hmC-independent processes are implicated in active DNA demethylation in bovine embryos. The recruitment of XRCC1 to damaged paternal pronuclei indicates that oxidative DNA lesions drive BER to repair DNA at the expense of DNA demethylation. Finally, this study highlighted striking differences in DNA methylation dynamics between bovine and mouse zygotes that will facilitate the understanding of the dynamics of DNA methylation in early development. CONCLUSIONS: The data demonstrate that oxidative stress in sperm has an impact not only on DNA integrity but also on the dynamics of epigenetic reprogramming, which may harm the paternal genetic and epigenetic contribution to the developing embryo and affect embryo development and embryo quality.</p>',
'date' => '2018-10-17',
'pmid' => 'http://www.pubmed.gov/30333056',
'doi' => '10.1186/s13072-018-0224-y',
'modified' => '2018-11-09 11:10:58',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 40 => array(
'id' => '3404',
'name' => 'Integrated analysis of DNA methylation profiling and gene expression profiling identifies novel markers in lung cancer in Xuanwei, China.',
'authors' => 'Wang J, Duan Y, Meng QH, Gong R, Guo C, Zhao Y, Zhang Y',
'description' => '<p>BACKGROUND: Aberrant DNA methylation occurs frequently in cancer. The aim of this study was to identify novel methylation markers in lung cancer in Xuanwei, China, through integrated genome-wide DNA methylation and gene expression studies. METHODS: Differentially methylated regions (DMRs) and differentially expressed genes (DEGs) were detected on 10 paired lung cancer tissues and noncancerous lung tissues by methylated DNA immunoprecipitation combined with microarray (MeDIP-chip) and gene expression microarray analyses, respectively. Integrated analysis of DMRs and DEGs was performed to screen out candidate methylation-related genes. Both methylation and expression changes of the candidate genes were further validated and analyzed. RESULTS: Compared with normal lung tissues, lung cancer tissues expressed a total of 6,899 DMRs, including 5,788 hypermethylated regions and 1,111 hypomethylated regions. Integrated analysis of DMRs and DEGs identified 45 tumor-specific candidate genes: 38 genes whose DMRs were hypermethylated and expression was downregulated, and 7 genes whose DMRs were hypomethylated and expression was upregulated. The methylation and expression validation results identified 4 candidate genes (STXBP6, BCL6B, FZD10, and HSPB6) that were significantly hypermethylated and downregulated in most of the tumor tissues compared with the noncancerous lung tissues. CONCLUSIONS: This integrated analysis of genome-wide DNA methylation and gene expression in lung cancer in Xuanwei revealed several genes regulated by promoter methylation that have not been described in lung cancer before. These results provide new insight into the carcinogenesis of lung cancer in Xuanwei and represent promising new diagnostic and therapeutic targets.</p>',
'date' => '2018-10-04',
'pmid' => 'http://www.pubmed.gov/30286088',
'doi' => '10.1371/journal.pone.0203155',
'modified' => '2018-11-09 11:24:21',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 41 => array(
'id' => '3417',
'name' => 'mGlu1 Receptors Monopolize the Synaptic Control of Cerebellar Purkinje Cells by Epigenetically Down-Regulating mGlu5 Receptors.',
'authors' => 'Notartomaso S, Nakao H, Mascio G, Scarselli P, Cannella M, Zappulla C, Madonna M, Motolese M, Gradini R, Liberatore F, Zonta M, Carmignoto G, Battaglia G, Bruno V, Watanabe M, Aiba A, Nicoletti F',
'description' => '<p>In cerebellar Purkinje cells (PCs) type-1 metabotropic glutamate (mGlu1) receptors play a key role in motor learning and drive the refinement of synaptic innervation during postnatal development. The cognate mGlu5 receptor is absent in mature PCs and shows low expression levels in the adult cerebellar cortex. Here we found that mGlu5 receptors were heavily expressed by PCs in the early postnatal life, when mGlu1α receptors were barely detectable. The developmental decline of mGlu5 receptors coincided with the appearance of mGlu1α receptors in PCs, and both processes were associated with specular changes in CpG methylation in the corresponding gene promoters. It was the mGlu1 receptor that drove the elimination of mGlu5 receptors from PCs, as shown by data obtained with conditional mGlu1α receptor knockout mice and with targeted pharmacological treatments during critical developmental time windows. The suppressing activity of mGlu1 receptors on mGlu5 receptor was maintained in mature PCs, suggesting that expression of mGlu1α and mGlu5 receptors is mutually exclusive in PCs. These findings add complexity to the the finely tuned mechanisms that regulate PC biology during development and in the adult life and lay the groundwork for an in-depth analysis of the role played by mGlu5 receptors in PC maturation.</p>',
'date' => '2018-09-06',
'pmid' => 'http://www.pubmed.gov/30190524',
'doi' => '10.1038/s41598-018-31369-7',
'modified' => '2018-12-31 11:36:04',
'created' => '2018-12-04 09:51:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 42 => array(
'id' => '3640',
'name' => 'Determination of the presence of 5-methylcytosine in Paramecium tetraurelia.',
'authors' => 'Singh A, Vancura A, Woycicki RK, Hogan DJ, Hendrick AG, Nowacki M',
'description' => '<p>5-methylcytosine DNA methylation regulates gene expression and developmental programming in a broad range of eukaryotes. However, its presence and potential roles in ciliates, complex single-celled eukaryotes with germline-somatic genome specialization via nuclear dimorphism, are largely uncharted. While canonical cytosine methyltransferases have not been discovered in published ciliate genomes, recent studies performed in the stichotrichous ciliate Oxytricha trifallax suggest de novo cytosine methylation during macronuclear development. In this study, we applied bisulfite genome sequencing, DNA mass spectrometry and antibody-based fluorescence detection to investigate the presence of DNA methylation in Paramecium tetraurelia. While the antibody-based methods suggest cytosine methylation, DNA mass spectrometry and bisulfite sequencing reveal that levels are actually below the limit of detection. Our results suggest that Paramecium does not utilize 5-methylcytosine DNA methylation as an integral part of its epigenetic arsenal.</p>',
'date' => '2018-06-07',
'pmid' => 'http://www.pubmed.gov/30379964',
'doi' => '10.1371/journal.',
'modified' => '2019-06-07 10:22:28',
'created' => '2019-06-06 12:11:18',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 43 => array(
'id' => '3458',
'name' => 'Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.',
'authors' => 'Li T, Wang L, Du Y, Xie S, Yang X, Lian F, Zhou Z, Qian C',
'description' => '<p>UHRF1 plays multiple roles in regulating DNMT1-mediated DNA methylation maintenance during DNA replication. The UHRF1 C-terminal RING finger functions as an ubiquitin E3 ligase to establish histone H3 ubiquitination at Lys18 and/or Lys23, which is subsequently recognized by DNMT1 to promote its localization onto replication foci. Here, we present the crystal structure of DNMT1 RFTS domain in complex with ubiquitin and highlight a unique ubiquitin binding mode for the RFTS domain. We provide evidence that UHRF1 N-terminal ubiquitin-like domain (UBL) also binds directly to DNMT1. Despite sharing a high degree of structural similarity, UHRF1 UBL and ubiquitin bind to DNMT1 in a very distinct fashion and exert different impacts on DNMT1 enzymatic activity. We further show that the UHRF1 UBL-mediated interaction between UHRF1 and DNMT1, and the binding of DNMT1 to ubiquitinated histone H3 that is catalyzed by UHRF1 RING domain are critical for the proper subnuclear localization of DNMT1 and maintenance of DNA methylation. Collectively, our study adds another layer of complexity to the regulatory mechanism of DNMT1 activation by UHRF1 and supports that individual domains of UHRF1 participate and act in concert to maintain DNA methylation patterns.</p>',
'date' => '2018-04-06',
'pmid' => 'http://www.pubmed.gov/29471350',
'doi' => '10.1093/nar/gky104',
'modified' => '2019-02-15 21:14:42',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 44 => array(
'id' => '3475',
'name' => 'Epigenetics and early domestication: differences in hypothalamic DNA methylation between red junglefowl divergently selected for high or low fear of humans.',
'authors' => 'Bélteky J, Agnvall B, Bektic L, Höglund A, Jensen P, Guerrero-Bosagna C',
'description' => '<p>BACKGROUND: Domestication of animals leads to large phenotypic alterations within a short evolutionary time-period. Such alterations are caused by genomic variations, yet the prevalence of modified traits is higher than expected if they were caused only by classical genetics and mutations. Epigenetic mechanisms may also be important in driving domesticated phenotypes such as behavior traits. Gene expression can be modulated epigenetically by mechanisms such as DNA methylation, resulting in modifications that are not only variable and susceptible to environmental stimuli, but also sometimes transgenerationally stable. To study such mechanisms in early domestication, we used as model two selected lines of red junglefowl (ancestors of modern chickens) that were bred for either high or low fear of humans over five generations, and investigated differences in hypothalamic DNA methylation between the two populations. RESULTS: Twenty-two 1-kb windows were differentially methylated between the two selected lines at p < 0.05 after false discovery rate correction. The annotated functions of the genes within these windows indicated epigenetic regulation of metabolic and signaling pathways, which agrees with the changes in gene expression that were previously reported for the same tissue and animals. CONCLUSIONS: Our results show that selection for an important domestication-related behavioral trait such as tameness can cause divergent epigenetic patterns within only five generations, and that these changes could have an important role in chicken domestication.</p>',
'date' => '2018-04-02',
'pmid' => 'http://www.pubmed.gov/29609558',
'doi' => '10.1186/s12711-018-0384-z',
'modified' => '2019-02-15 20:32:37',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 45 => array(
'id' => '3393',
'name' => 'Copper induces expression and methylation changes of early development genes in Crassostrea gigas embryos.',
'authors' => 'Sussarellu R, Lebreton M, Rouxel J, Akcha F, Rivière G',
'description' => '<p>Copper contamination is widespread along coastal areas and exerts adverse effects on marine organisms such as mollusks. In the Pacific oyster, copper induces severe developmental abnormalities during early life stages; however, the underlying molecular mechanisms are largely unknown. This study aims to better understand whether the embryotoxic effects of copper in Crassostrea gigas could be mediated by alterations in gene expression, and the putative role of DNA methylation, which is known to contribute to gene regulation in early embryo development. For that purpose, oyster embryos were exposed to 4 nominal copper concentrations (0.1, 1, 10 and 20 μg L Cu) during early development assays. Embryotoxicity was monitored through the oyster embryo-larval bioassay at the D-larva stage 24 h post fertilization (hpf) and genotoxicity at gastrulation 7 hpf. In parallel, the relative expression of 15 genes encoding putative homeotic, biomineralization and DNA methylation proteins was measured at three developmental stages (3 hpf morula stage, 7 hpf gastrula stage, 24 hpf D-larvae stage) using RT-qPCR. Global DNA content in methylcytosine and hydroxymethylcytosine were measured by HPLC and gene-specific DNA methylation levels were monitored using MeDIP-qPCR. A significant increase in larval abnormalities was observed from copper concentrations of 10 μg L, while significant genotoxic effects were detected at 1 μg L and above. All the selected genes presented a stage-dependent expression pattern, which was impaired for some homeobox and DNA methylation genes (Notochord, HOXA1, HOX2, Lox5, DNMT3b and CXXC-1) after copper exposure. While global DNA methylation (5-methylcytosine) at gastrula stage didn't show significant changes between experimental conditions, 5-hydroxymethylcytosine, its degradation product, decreased upon copper treatment. The DNA methylation of exons and the transcript levels were correlated in control samples for HOXA1 but such a correlation was diminished following copper exposure. The methylation level of some specific gene regions (HoxA1, Hox2, Engrailed2 and Notochord) displayed changes upon copper exposure. Such changes were gene and exon-specific and no obvious global trends could be identified. Our study suggests that the embryotoxic effects of copper in oysters could involve homeotic gene expression impairment possibly by changing DNA methylation levels.</p>',
'date' => '2018-03-01',
'pmid' => 'http://www.pubmed.gov/29353135',
'doi' => '10.1016/j.aquatox.2018.01.001',
'modified' => '2018-11-09 12:21:38',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 46 => array(
'id' => '3448',
'name' => 'Aberrant methylated key genes of methyl group metabolism within the molecular etiology of urothelial carcinogenesis.',
'authors' => 'Erichsen L, Ghanjati F, Beermann A, Poyet C, Hermanns T, Schulz WA, Seifert HH, Wild PJ, Buser L, Kröning A, Braunstein S, Anlauf M, Jankowiak S, Hassan M, Bendhack ML, Araúzo-Bravo MJ, Santourlidis S',
'description' => '<p>Urothelial carcinoma (UC), the most common cancer of the urinary bladder causes severe morbidity and mortality, e.g. about 40.000 deaths in the EU annually, and incurs considerable costs for the health system due to the need for prolonged treatments and long-term monitoring. Extensive aberrant DNA methylation is described to prevail in urothelial carcinoma and is thought to contribute to genetic instability, altered gene expression and tumor progression. However, it is unknown how this epigenetic alteration arises during carcinogenesis. Intact methyl group metabolism is required to ensure maintenance of cell-type specific methylomes and thereby genetic integrity and proper cellular function. Here, using two independent techniques for detecting DNA methylation, we observed DNA hypermethylation of the 5'-regulatory regions of the key methyl group metabolism genes ODC1, AHCY and MTHFR in early urothelial carcinoma. These hypermethylation events are associated with genome-wide DNA hypomethylation which is commonly associated with genetic instability. We therefore infer that hypermethylation of methyl group metabolism genes acts in a feed-forward cycle to promote additional DNA methylation changes and suggest a new hypothesis on the molecular etiology of urothelial carcinoma.</p>',
'date' => '2018-02-22',
'pmid' => 'http://www.pubmed.gov/29472622',
'doi' => '10.1038/s41598-018-21932-7',
'modified' => '2019-02-15 21:31:04',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 47 => array(
'id' => '3383',
'name' => 'Genome-wide analysis of day/night DNA methylation differences in Populus nigra.',
'authors' => 'Ding C.J. et al.',
'description' => '<p>DNA methylation is an important mechanism of epigenetic modification. Methylation changes during stress responses and developmental processes have been well studied; however, their role in plant adaptation to the day/night cycle is poorly understood. In this study, we detected global methylation patterns in leaves of the black poplar Populus nigra 'N46' at 8:00 and 24:00 by methylated DNA immunoprecipitation sequencing (MeDIP-seq). We found 10,027 and 10,242 genes to be methylated in the 8:00 and 24:00 samples, respectively. The methylated genes appeared to be involved in multiple biological processes, molecular functions, and cellular components, suggesting important roles for DNA methylation in poplar cells. Comparing the 8:00 and 24:00 samples, only 440 differentially methylated regions (DMRs) overlapped with genic regions, including 193 hyper- and 247 hypo-methylated DMRs, and may influence the expression of 137 downstream genes. Most hyper-methylated genes were associated with transferase activity, kinase activity, and phosphotransferase activity, whereas most hypo-methylated genes were associated with protein binding, ATP binding, and adenyl ribonucleotide binding, suggesting that different biological processes were activated during the day and night. Our results indicated that methylated genes were prevalent in the poplar genome, but that only a few of these participated in diurnal gene expression regulation.</p>',
'date' => '2018-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29293569',
'doi' => '',
'modified' => '2018-08-07 09:45:38',
'created' => '2018-08-07 09:45:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 48 => array(
'id' => '3384',
'name' => 'Obligatory and facilitative allelic variation in the DNA methylome within common disease-associated loci',
'authors' => 'Bell C.G. et al.',
'description' => '<p>Integrating epigenetic data with genome-wide association study (GWAS) results can reveal disease mechanisms. The genome sequence itself also shapes the epigenome, with CpG density and transcription factor binding sites (TFBSs) strongly encoding the DNA methylome. Therefore, genetic polymorphism impacts on the observed epigenome. Furthermore, large genetic variants alter epigenetic signal dosage. Here, we identify DNA methylation variability between GWAS-SNP risk and non-risk haplotypes. In three subsets comprising 3128 MeDIP-seq peripheral-blood DNA methylomes, we find 7173 consistent and functionally enriched Differentially Methylated Regions. 36.8% can be attributed to common non-SNP genetic variants. CpG-SNPs, as well as facilitative TFBS-motifs, are also enriched. Highlighting their functional potential, CpG-SNPs strongly associate with allele-specific DNase-I hypersensitivity sites. Our results demonstrate strong DNA methylation allelic differences driven by obligatory or facilitative genetic effects, with potential direct or regional disease-related repercussions. These allelic variations require disentangling from pure tissue-specific modifications, may influence array studies, and imply underestimated population variability in current reference epigenomes.</p>',
'date' => '2018-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29295990',
'doi' => '',
'modified' => '2018-08-07 10:13:12',
'created' => '2018-08-07 10:13:12',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 49 => array(
'id' => '3508',
'name' => 'Analysis of DNA methylome and transcriptome profiling following Gibberellin A3 (GA3) foliar application in Nicotiana tabacum L.',
'authors' => 'Manoharlal Raman, Saiprasad G. V. S., Kaikala Vinay, Suresh Kumar R., Kovařík Ales',
'description' => '<p>The present work investigated a comprehensive genome-wide landscape of DNA methylome and its relationship with transcriptome upon gibberellin A3 (GA3) foliar application under practical field conditions in solanaceae model, Nicotiana tabacum L. Methylated DNA Immunoprecipitation-Sequencing (MeDIP-Seq) analysis uncovered over 82% (18,456) of differential methylated regions (DMRs) in intergenic-region. Within protein-coding region, 2339 and 1685 of identified DMRs were observed in genebody- and promoter-region, respectively. Microarray study revealed 7032 differential expressed genes (DEGs) with 3507 and 3525 genes displaying upand down-regulation, respectively. Integration analysis revealed 520 unique non-redundant annotated DMRs overlapping with DEGs. Our results indicated that GA3 induced DNA hypo- as well as hyper-methylation were associated with both gene-silencing and -activation. No complete biasness or correlation was observed in either of the promoter- or genebody-regions, which otherwise showed an overall trend towards GA3 induced global DNA hypo-methylation. Taken together, our results suggested that differential DNA methylation mediated by GA3 may only play a permissive role in regulating the gene expression.</p>',
'date' => '2018-01-01',
'pmid' => 'https://link.springer.com/article/10.1007/s40502-018-0393-5',
'doi' => '10.1007/s40502-018-0393-5',
'modified' => '2022-05-18 18:43:47',
'created' => '2019-02-27 12:54:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 50 => array(
'id' => '3297',
'name' => '5-Methylcytosine RNA Methylation in Arabidopsis Thaliana',
'authors' => 'Cui X. et al.',
'description' => '<p>5-Methylcytosine (m<sup>5</sup>C) is a well-characterized DNA modification, and is also predominantly reported in abundant non-coding RNAs in both prokaryotes and eukaryotes. However, the distribution and biological functions of m<sup>5</sup>C in plant mRNAs remain largely unknown. Here, we report transcriptome-wide profiling of RNA m<sup>5</sup>C in Arabidopsis thaliana by applying m<sup>5</sup>C RNA immunoprecipitation followed by a deep-sequencing approach (m<sup>5</sup>C-RIP-seq). LC-MS/MS and dot blot analyses reveal a dynamic pattern of m<sup>5</sup>C mRNA modification in various tissues and at different developmental stages. m<sup>5</sup>C-RIP-seq analysis identified 6045 m<sup>5</sup>C peaks in 4465 expressed genes in young seedlings. We found that m<sup>5</sup>C is enriched in coding sequences with two peaks located immediately after start codons and before stop codons, and is associated with mRNAs with low translation activity. We further demonstrated that an RNA (cytosine-5)-methyltransferase, tRNA-specific methyltransferase 4B (TRM4B), exhibits m<sup>5</sup>C RNA methyltransferase activity. Mutations in TRM4B display defects in root development and decreased m<sup>5</sup>C peaks. TRM4B affects the transcript levels of the genes involved in root development, which is positively correlated with their mRNA stability and m<sup>5</sup>C levels. Our results suggest that m<sup>5</sup>C in mRNA is a new epitranscriptome marker inArabidopsis, and that regulation of this modification is an integral part of gene regulatory networks underlying plant development.</p>',
'date' => '2017-11-06',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28965832',
'doi' => '',
'modified' => '2017-12-04 11:10:34',
'created' => '2017-12-04 11:10:34',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 51 => array(
'id' => '3220',
'name' => 'Maternal obesity programs increased leptin gene expression in rat male offspring via epigenetic modifications in a depot-specific manner',
'authors' => 'Lecoutre S. et al.',
'description' => '<div class="">
<h4>OBJECTIVE:</h4>
<p><abstracttext label="OBJECTIVE" nlmcategory="OBJECTIVE">According to the Developmental Origin of Health and Disease (DOHaD) concept, maternal obesity and accelerated growth in neonates predispose offspring to white adipose tissue (WAT) accumulation. In rodents, adipogenesis mainly develops during lactation. The mechanisms underlying the phenomenon known as developmental programming remain elusive. We previously reported that adult rat offspring from high-fat diet-fed dams (called HF) exhibited hypertrophic adipocyte, hyperleptinemia and increased leptin mRNA levels in a depot-specific manner. We hypothesized that leptin upregulation occurs via epigenetic malprogramming, which takes place early during development of WAT.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">As a first step, we identified <i>in silico</i> two potential enhancers located upstream and downstream of the leptin transcription start site that exhibit strong dynamic epigenomic remodeling during adipocyte differentiation. We then focused on epigenetic modifications (methylation, hydroxymethylation, and histone modifications) of the promoter and the two potential enhancers regulating leptin gene expression in perirenal (pWAT) and inguinal (iWAT) fat pads of HF offspring during lactation (postnatal days 12 (PND12) and 21 (PND21)) and in adulthood.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">PND12 is an active period for epigenomic remodeling in both deposits especially in the upstream enhancer, consistent with leptin gene induction during adipogenesis. Unlike iWAT, some of these epigenetic marks were still observable in pWAT of weaned HF offspring. Retained marks were only visible in pWAT of 9-month-old HF rats that showed a persistent "expandable" phenotype.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Consistent with the DOHaD hypothesis, persistent epigenetic remodeling occurs at regulatory regions especially within intergenic sequences, linked to higher leptin gene expression in adult HF offspring in a depot-specific manner.</abstracttext></p>
</div>',
'date' => '2017-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5518658/',
'doi' => '',
'modified' => '2017-08-18 13:56:40',
'created' => '2017-08-18 13:56:40',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 52 => array(
'id' => '3204',
'name' => 'Increased 5-hydroxymethylation levels in the hippocampus of rat extinguished from cocaine self-administration',
'authors' => 'Sadakierska-Chudy A. et al.',
'description' => '<p>Drug craving and relapse risk during abstinence from cocaine are thought to be caused by persistent changes in transcription and chromatin regulation. Although several brain regions are involved in these processes, the hippocampus seems to play an important role in context-evoked craving and drug-seeking behavior. Only a few studies have examined epigenetic alterations during a period of cocaine abstinence. To investigate the effects of cocaine abstinence on DNA methylation and gene expression, rats that self-administered the drug underwent cocaine abstinence in two time points with extinction training. During the cocaine extinction, we observed elevated global 5-hydroxymethylcytosine(5-hmC) levels with a concurrent increase in Tet3 transcript levels. Moreover, we did not find significant alterations in the levels of Tet3 mRNA and 5-hmC in rats subjected to cocaine abstinence without extinction training. Additionally, our findings demonstrated that the expression of Tet3 target genes was activated. Besides, altered DNA methylation was detected at promoter regions of miRNAs, such as miR-30d and miR-let7i. Further in silico analysis provided evidence that these two molecules targeted the 3' UTR region of the Tet3 gene and thus may contribute to its post-transcriptional regulation. This study has presented novel findings in the hippocampus of rats that underwent extinction training following cocaine self-administration. The alterations in the Tet3 gene expression and the level of 5-hmC may play an important role in extinction learning and the reduction of subsequent cocaine seeking.</p>',
'date' => '2017-07-27',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28422379',
'doi' => '',
'modified' => '2017-07-03 10:21:48',
'created' => '2017-07-03 10:21:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 53 => array(
'id' => '3225',
'name' => 'The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation',
'authors' => 'Leone S. et al.',
'description' => '<p>Long non-coding RNAs (lncRNAs) have been implicated in the regulation of chromatin conformation and epigenetic patterns. lncRNA expression levels are widely taken as an indicator for functional properties. However, the role of RNA processing in modulating distinct features of the same lncRNA is less understood. The establishment of heterochromatin at rRNA genes depends on the processing of IGS-rRNA into pRNA, a reaction that is impaired in embryonic stem cells (ESCs) and activated only upon differentiation. The production of mature pRNA is essential since it guides the repressor TIP5 to rRNA genes, and IGS-rRNA abolishes this process. Through screening for IGS-rRNA-binding proteins, we here identify the RNA helicase DHX9 as a regulator of pRNA processing. DHX9 binds to rRNA genes only upon ESC differentiation and its activity guides TIP5 to rRNA genes and establishes heterochromatin. Remarkably, ESCs depleted of DHX9 are unable to differentiate and this phenotype is reverted by the addition of pRNA, whereas providing IGS-rRNA and pRNA mutants deficient for TIP5 binding are not sufficient. Our results reveal insights into lncRNA biogenesis during development and support a model in which the state of rRNA gene chromatin is part of the regulatory network that controls exit from pluripotency and initiation of differentiation pathways.</p>',
'date' => '2017-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28588071',
'doi' => '',
'modified' => '2017-08-22 13:52:28',
'created' => '2017-08-22 13:52:28',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 54 => array(
'id' => '3233',
'name' => 'Pramel7 mediates ground-state pluripotency through proteasomal-epigenetic combined pathways.',
'authors' => 'Graf U. et al.',
'description' => '<p>Naive pluripotency is established in preimplantation epiblast. Embryonic stem cells (ESCs) represent the immortalization of naive pluripotency. 2i culture has optimized this state, leading to a gene signature and DNA hypomethylation closely comparable to preimplantation epiblast, the developmental ground state. Here we show that Pramel7 (PRAME-like 7), a protein highly expressed in the inner cell mass (ICM) but expressed at low levels in ESCs, targets for proteasomal degradation UHRF1, a key factor for DNA methylation maintenance. Increasing Pramel7 expression in serum-cultured ESCs promotes a preimplantation epiblast-like gene signature, reduces UHRF1 levels and causes global DNA hypomethylation. Pramel7 is required for blastocyst formation and its forced expression locks ESCs in pluripotency. Pramel7/UHRF1 expression is mutually exclusive in ICMs whereas Pramel7-knockout embryos express high levels of UHRF1. Our data reveal an as-yet-unappreciated dynamic nature of DNA methylation through proteasome pathways and offer insights that might help to improve ESC culture to reproduce in vitro the in vivo ground-state pluripotency.</p>',
'date' => '2017-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28604677',
'doi' => '',
'modified' => '2017-08-24 09:57:28',
'created' => '2017-08-24 09:57:28',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 55 => array(
'id' => '3200',
'name' => 'CHD4 Has Oncogenic Functions in Initiating and Maintaining Epigenetic Suppression of Multiple Tumor Suppressor Genes',
'authors' => 'Xia L. et al.',
'description' => '<p>An oncogenic role for CHD4, a NuRD component, is defined for initiating and supporting tumor suppressor gene (TSG) silencing in human colorectal cancer. CHD4 recruits repressive chromatin proteins to sites of DNA damage repair, including DNA methyltransferases where it imposes de novo DNA methylation. At TSGs, CHD4 retention helps maintain DNA hypermethylation-associated transcriptional silencing. CHD4 is recruited by the excision repair protein OGG1 for oxidative damage to interact with the damage-induced base 8-hydroxydeoxyguanosine (8-OHdG), while ZMYND8 recruits it to double-strand breaks. CHD4 knockdown activates silenced TSGs, revealing their role for blunting colorectal cancer cell proliferation, invasion, and metastases. High CHD4 and 8-OHdG levels plus low expression of TSGs strongly correlates with early disease recurrence and decreased overall survival.</p>',
'date' => '2017-05-08',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28486105',
'doi' => '',
'modified' => '2017-07-03 09:56:32',
'created' => '2017-07-03 09:56:32',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 56 => array(
'id' => '3170',
'name' => 'Critical threshold levels of DNA methyltransferase 1 are required to maintain DNA methylation across the genome in human cancer cells.',
'authors' => 'Cai Y. et al.',
'description' => '<p>Reversing DNA methylation abnormalities and associated gene silencing, through inhibiting DNA methyltransferases (DNMTs) is an important potential cancer therapy paradigm. Maximizing this potential requires defining precisely how these enzymes maintain genome-wide, cancer-specific DNA methylation. To date, there is incomplete understanding of precisely how the three DNMTs, 1, 3A, and 3B, interact for maintaining DNA methylation abnormalities in cancer. By combining genetic and shRNA depletion strategies, we define not only a dominant role for DNA methyltransferase 1 (DNMT1) but also distinct roles of 3A and 3B in genome-wide DNA methylation maintenance. Lowering DNMT1 below a threshold level is required for maximal loss of DNA methylation at all genomic regions, including gene body and enhancer regions, and for maximally reversing abnormal promoter DNA hypermethylation and associated gene silencing to reexpress key genes. It is difficult to reach this threshold with patient-tolerable doses of current DNMT inhibitors (DNMTIs). We show that new approaches, like decreasing the DNMT targeting protein, UHRF1, can augment the DNA demethylation capacities of existing DNA methylation inhibitors for fully realizing their therapeutic potential.</p>',
'date' => '2017-04-27',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28232479',
'doi' => '',
'modified' => '2017-05-10 15:31:12',
'created' => '2017-05-10 15:31:12',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 57 => array(
'id' => '3228',
'name' => 'Regulation of DNA demethylation by the XPC DNA repair complex in somatic and pluripotent stem cells.',
'authors' => 'Ho J.J. et al.',
'description' => '<p>Faithful resetting of the epigenetic memory of a somatic cell to a pluripotent state during cellular reprogramming requires DNA methylation to silence somatic gene expression and dynamic DNA demethylation to activate pluripotency gene transcription. The removal of methylated cytosines requires the base excision repair enzyme TDG, but the mechanism by which TDG-dependent DNA demethylation occurs in a rapid and site-specific manner remains unclear. Here we show that the XPC DNA repair complex is a potent accelerator of global and locus-specific DNA demethylation in somatic and pluripotent stem cells. XPC cooperates with TDG genome-wide to stimulate the turnover of essential intermediates by overcoming slow TDG-abasic product dissociation during active DNA demethylation. We further establish that DNA demethylation induced by XPC expression in somatic cells overcomes an early epigenetic barrier in cellular reprogramming and facilitates the generation of more robust induced pluripotent stem cells, characterized by enhanced pluripotency-associated gene expression and self-renewal capacity. Taken together with our previous studies establishing the XPC complex as a transcriptional coactivator, our findings underscore two distinct but complementary mechanisms by which XPC influences gene regulation by coordinating efficient TDG-mediated DNA demethylation along with active transcription during somatic cell reprogramming.</p>',
'date' => '2017-04-05',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28512237',
'doi' => '',
'modified' => '2017-08-23 14:20:13',
'created' => '2017-08-23 14:20:13',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 58 => array(
'id' => '3142',
'name' => 'Epigenetic regulation of RELN and GAD1 in the frontal cortex (FC) of autism spectrum disorder (ASD) subjects',
'authors' => 'Zhubi A. et al.',
'description' => '<p>Both Reelin (RELN) and glutamate decarboxylase 67 (GAD1) have been implicated in the pathophysiology of Autism Spectrum Disorders (ASD). We have previously shown that both mRNAs are reduced in the cerebella (CB) of ASD subjects through a mechanism that involves increases in the amounts of MECP2 binding to the corresponding promoters. In the current study, we examined the expression of RELN, GAD1, GAD2, and several other mRNAs implicated in this disorder in the frontal cortices (FC) of ASD and CON subjects. We also focused on the role that epigenetic processes play in the regulation of these genes in ASD brain. Our goal is to better understand the molecular basis for the down-regulation of genes expressed in GABAergic neurons in ASD brains. We measured mRNA levels corresponding to selected GABAergic genes using qRT-PCR in RNA isolated from both ASD and CON groups. We determined the extent of binding of MECP2 and DNMT1 repressor proteins by chromatin immunoprecipitation (ChIP) assays. The amount of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) present in the promoters of the target genes was quantified by methyl DNA immunoprecipitation (MeDIP) and hydroxyl MeDIP (hMeDIP). We detected significant reductions in the mRNAs associated with RELN and GAD1 and significant increases in mRNAs encoding the Ten-eleven Translocation (TET) enzymes 1, 2, and 3. We also detected increased MECP2 and DNMT1 binding to the corresponding promoter regions of GAD1, RELN, and GAD2. Interestingly, there were decreased amounts of 5mC at both promoters and little change in 5hmC content in these same DNA fragments. Our data demonstrate that RELN, GAD1, and several other genes selectively expressed in GABAergic neurons, are down-regulated in post-mortem ASD FC. In addition, we observed increased DNMT1 and MECP2 binding at the corresponding promoters of these genes. The finding of increased MECP2 binding to the RELN, GAD1 and GAD2 promoters, with reduced amounts of 5mC and unchanged amounts of 5hmC present in these regions, suggests the possibility that DNMT1 interacts with and alters MECP2 binding properties to selected promoters. Comparisons between data obtained from the FC with CB studies showed some common themes between brain regions which are discussed.</p>',
'date' => '2017-02-14',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28229923',
'doi' => '',
'modified' => '2017-03-23 14:58:21',
'created' => '2017-03-23 14:58:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 59 => array(
'id' => '3126',
'name' => 'Intergenerational Transmission of Enhanced Seizure Susceptibility after Febrile Seizures',
'authors' => 'Wu D. et al.',
'description' => '<p>Environmental exposure early in development plays a role in susceptibility to disease in later life. Here, we demonstrate that prolonged febrile seizures induced by exposure of rat pups to a hyperthermic environment enhance seizure susceptibility not only in these hyperthermia-treated rats but also in their future offspring, even if the offspring never experience febrile seizures. This transgenerational transmission was intensity-dependent and was mainly from mothers to their offspring. The transmission was associated with DNA methylation. Thus, our study supports a “Lamarckian”-like mechanism of pathogenesis and the crucial role of epigenetic factors in neurological conditions.</p>',
'date' => '2017-02-08',
'pmid' => 'http://www.sciencedirect.com/science/article/pii/S2352396417300658',
'doi' => '',
'modified' => '2017-02-23 11:05:25',
'created' => '2017-02-23 11:05:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 60 => array(
'id' => '3125',
'name' => 'Pharmacological inhibition of DNA methyltransferase 1 promotes neuronal differentiation from rodent and human nasal olfactory stem/progenitor cell cultures',
'authors' => 'Franco I. et al.',
'description' => '<p>Nasal olfactory stem and neural progenitor cells (NOS/PCs) are considered possible tools for regenerative stem cell therapies in neurodegenerative diseases. Neurogenesis is a complex process regulated by extrinsic and intrinsic signals that include DNA-methylation and other chromatin modifications that could be experimentally manipulated in order to increase neuronal differentiation. The aim of the present study was the characterization of primary cultures and consecutive passages (P2-P10) of NOS/PCs isolated from male Swiss-Webster (mNOS/PCs) or healthy humans (hNOS/PCs). We evaluated and compared cellular morphology, proliferation rates and the expression pattern of pluripotency-associated markers and DNA methylation-associated gene expression in these cultures. Neuronal differentiation was induced by exposure to all-trans retinoic acid and forskolin for 7 days and evaluated by morphological analysis and immunofluorescence against neuronal markers MAP2, NSE and MAP1B. In response to the inductive cues mNOS/PCs expressed NSE (75.67%) and MAP2 (35.34%); whereas the majority of the hNOS/PCs were immunopositive to MAP1B. Treatment with procainamide, a specific inhibitor of DNA methyltransferase 1 (DNMT1), increases in the number of forskolin‘/retinoic acid-induced mature neuronal marker-expressing mNOS/PCs cells and enhances neurite development in hNOS/PCs. Our results indicate that mice and human nasal olfactory stem/progenitors cells share pluripotency-related gene expression suggesting that their application for stem cell therapy is worth pursuing and that DNA methylation inhibitors could be efficient tools to enhance neuronal differentiation from these cells.</p>',
'date' => '2017-02-01',
'pmid' => 'http://www.sciencedirect.com/science/article/pii/S0736574816303665',
'doi' => '',
'modified' => '2017-02-16 10:34:07',
'created' => '2017-02-16 10:34:07',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 61 => array(
'id' => '3119',
'name' => 'Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain',
'authors' => 'Amort T. et al.',
'description' => '<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec1">
<h3 xmlns="" class="Heading">Background</h3>
<p id="Par1" class="Para">Recent work has identified and mapped a range of posttranscriptional modifications in mRNA, including methylation of the N6 and N1 positions in adenine, pseudouridylation, and methylation of carbon 5 in cytosine (m5C). However, knowledge about the prevalence and transcriptome-wide distribution of m5C is still extremely limited; thus, studies in different cell types, tissues, and organisms are needed to gain insight into possible functions of this modification and implications for other regulatory processes.</p>
</div>
<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec2">
<h3 xmlns="" class="Heading">Results</h3>
<p id="Par2" class="Para">We have carried out an unbiased global analysis of m5C in total and nuclear poly(A) RNA of mouse embryonic stem cells and murine brain. We show that there are intriguing differences in these samples and cell compartments with respect to the degree of methylation, functional classification of methylated transcripts, and position bias within the transcript. Specifically, we observe a pronounced accumulation of m5C sites in the vicinity of the translational start codon, depletion in coding sequences, and mixed patterns of enrichment in the 3′ UTR. Degree and pattern of methylation distinguish transcripts modified in both embryonic stem cells and brain from those methylated in either one of the samples. We also analyze potential correlations between m5C and micro RNA target sites, binding sites of RNA binding proteins, and <em xmlns="" class="EmphasisTypeItalic">N</em>6-methyladenosine.</p>
</div>
<div xmlns="http://www.w3.org/1999/xhtml" class="AbstractSection" id="ASec3">
<h3 xmlns="" class="Heading">Conclusion</h3>
<p id="Par3" class="Para">Our study presents the first comprehensive picture of cytosine methylation in the epitranscriptome of pluripotent and differentiated stages in the mouse. These data provide an invaluable resource for future studies of function and biological significance of m5C in mRNA in mammals.</p>
</div>',
'date' => '2017-01-05',
'pmid' => 'https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1139-1',
'doi' => '',
'modified' => '2017-02-14 17:20:08',
'created' => '2017-02-14 17:20:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 62 => array(
'id' => '3061',
'name' => 'Novel regional age-associated DNA methylation changes within human common disease-associated loci',
'authors' => 'Bell CG et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">Advancing age progressively impacts on risk and severity of chronic disease. It also modifies the epigenome, with changes in DNA methylation, due to both random drift and variation within specific functional loci.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">In a discovery set of 2238 peripheral-blood genome-wide DNA methylomes aged 19-82 years, we identify 71 age-associated differentially methylated regions within the linkage disequilibrium blocks of the single nucleotide polymorphisms from the NIH genome-wide association study catalogue. This included 52 novel regions, 29 within loci not covered by 450 k or 27 k Illumina array, and with enrichment for DNase-I Hypersensitivity sites across the full range of tissues. These age-associated differentially methylated regions also show marked enrichment for enhancers and poised promoters across multiple cell types. In a replication set of 2084 DNA methylomes, 95.7 % of the age-associated differentially methylated regions showed the same direction of ageing effect, with 80.3 % and 53.5 % replicated to p < 0.05 and p < 1.85 × 10<sup>-8</sup>, respectively.</abstracttext></p>
<h4>CONCLUSION:</h4>
<p><abstracttext label="CONCLUSION" nlmcategory="CONCLUSIONS">By analysing the functionally enriched disease and trait-associated regions of the human genome, we identify novel epigenetic ageing changes, which could be useful biomarkers or provide mechanistic insights into age-related common diseases.</abstracttext></p>
</div>',
'date' => '2016-09-26',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27663977',
'doi' => '',
'modified' => '2016-11-04 10:56:10',
'created' => '2016-11-02 09:54:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 63 => array(
'id' => '3007',
'name' => '5-hydroxymethylcytosine marks postmitotic neural cells in the adult and developing vertebrate central nervous system',
'authors' => 'Diotel N et al.',
'description' => '<p>The epigenetic mark 5-hydroxymethylcytosine (5hmC) is a cytosine modification that is abundant in the central nervous system of mammals and which results from 5-methylcytosine oxidation by TET enzymes. Such a mark is suggested to play key roles in the regulation of chromatin structure and gene expression. However, its precise functions still remain poorly understood and information about its distribution in non-mammalian species is still lacking. Here, the distribution of 5hmC was investigated in the brain of adult zebrafish, African claw frog, and mouse in a comparative manner. We show that zebrafish neurons are endowed with high levels of 5hmC, whereas quiescent or proliferative neural progenitors show low to undetectable levels of the modified cytosine. In the brain of larval and juvenile Xenopus, 5hmC is also detected in neurons, while ventricular proliferative cells do not display this epigenetic mark. Similarly, 5hmC is enriched in neurons compared to neural progenitors of the ventricular zone in the mouse developing cortex. Interestingly, 5hmC colocalized with the methylated DNA binding protein MeCP2 and with the active chromatin histone modification H3K4me2 in mouse neurons. Taken together, our results show an evolutionarily conserved cerebral distribution of 5hmC between fish and tetrapods and reinforce the idea that 5hmC fulfills major functions in the control of chromatin activity in vertebrate neurons.</p>',
'date' => '2016-07-14',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27414756',
'doi' => '',
'modified' => '2016-08-29 09:24:44',
'created' => '2016-08-29 09:24:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 64 => array(
'id' => '2992',
'name' => 'Regulation of the DNA Methylation Landscape in Human Somatic Cell Reprogramming by the miR-29 Family',
'authors' => 'Hysolli E et al.',
'description' => 'Reprogramming to pluripotency after overexpression of OCT4, SOX2, KLF4, and MYC is accompanied by global genomic and epigenomic changes. Histone modification and DNA methylation states in induced pluripotent stem cells (iPSCs) have been shown to be highly similar to embryonic stem cells (ESCs). However, epigenetic differences still exist between iPSCs and ESCs. In particular, aberrant DNA methylation states found in iPSCs are a major concern when using iPSCs in a clinical setting. Thus, it is critical to find factors that regulate DNA methylation states in reprogramming. Here, we found that the miR-29 family is an important epigenetic regulator during human somatic cell reprogramming. Our global DNA methylation and hydroxymethylation analysis shows that DNA demethylation is a major event mediated by miR-29a depletion during early reprogramming, and that iPSCs derived from miR-29a depletion are epigenetically closer to ESCs. Our findings uncover an important miRNA-based approach to generate clinically robust iPSCs.',
'date' => '2016-07-12',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27373925',
'doi' => '10.1016/j.stemcr.2016.05.014',
'modified' => '2016-08-23 09:57:29',
'created' => '2016-08-23 09:57:29',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 65 => array(
'id' => '3011',
'name' => 'Genome-Wide DNA Methylation in Mixed Ancestry Individuals with Diabetes and Prediabetes from South Africa',
'authors' => 'Matsha TE et al.',
'description' => '<p>Aims. To conduct a genome-wide DNA methylation in individuals with type 2 diabetes, individuals with prediabetes, and control mixed ancestry individuals from South Africa. Methods. We used peripheral blood to perform genome-wide DNA methylation analysis in 3 individuals with screen detected diabetes, 3 individuals with prediabetes, and 3 individuals with normoglycaemia from the Bellville South Community, Cape Town, South Africa, who were age-, gender-, body mass index-, and duration of residency-matched. Methylated DNA immunoprecipitation (MeDIP) was performed by Arraystar Inc. (Rockville, MD, USA). Results. Hypermethylated DMRs were 1160 (81.97%) and 124 (43.20%), respectively, in individuals with diabetes and prediabetes when both were compared to subjects with normoglycaemia. Our data shows that genes related to the immune system, signal transduction, glucose transport, and pancreas development have altered DNA methylation in subjects with prediabetes and diabetes. Pathway analysis based on the functional analysis mapping of genes to KEGG pathways suggested that the linoleic acid metabolism and arachidonic acid metabolism pathways are hypomethylated in prediabetes and diabetes. Conclusions. Our study suggests that epigenetic changes are likely to be an early process that occurs before the onset of overt diabetes. Detailed analysis of DMRs that shows gradual methylation differences from control versus prediabetes to prediabetes versus diabetes in a larger sample size is required to confirm these findings.</p>',
'date' => '2016-06-28',
'pmid' => 'http://www.hindawi.com/journals/ije/2016/3172093/',
'doi' => '',
'modified' => '2016-08-29 10:27:14',
'created' => '2016-08-29 10:27:14',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 66 => array(
'id' => '2954',
'name' => 'Dnmt2/Trdmt1 as Mediator of RNA Polymerase II Transcriptional Activity in Cardiac Growth',
'authors' => 'Ghanbarian H et al.',
'description' => '<p>Dnmt2/Trdmt1 is a methyltransferase, which has been shown to methylate tRNAs. Deficient mutants were reported to exhibit various, seemingly unrelated, defects in development and RNA-mediated epigenetic heredity. Here we report a role in a distinct developmental regulation effected by a noncoding RNA. We show that Dnmt2-deficiency in mice results in cardiac hypertrophy. Echocardiographic measurements revealed that cardiac function is preserved notwithstanding the increased dimensions of the organ due to cardiomyocyte enlargement. Mechanistically, activation of the P-TEFb complex, a critical step for cardiac growth, results from increased dissociation of the negatively regulating Rn7sk non-coding RNA component in Dnmt2-deficient cells. Our data suggest that Dnmt2 plays an unexpected role for regulation of cardiac growth by modulating activity of the P-TEFb complex.</p>',
'date' => '2016-06-06',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27270731',
'doi' => ' 10.1371/journal.pone.0156953',
'modified' => '2016-06-14 15:49:17',
'created' => '2016-06-14 15:49:17',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 67 => array(
'id' => '2941',
'name' => 'Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers',
'authors' => 'Li L et al.',
'description' => '<p>Promoter CpG methylation is a fundamental regulatory process of gene expression. TET proteins are active CpG demethylases converting 5-methylcytosine to 5-hydroxymethylcytosine, with loss of 5 hmC as an epigenetic hallmark of cancers, indicating critical roles of TET proteins in epigenetic tumorigenesis. Through analysis of tumor methylomes, we discovered <i>TET1</i> as a methylated target, and further confirmed its frequent downregulation/methylation in cell lines and primary tumors of multiple carcinomas and lymphomas, including nasopharyngeal, esophageal, gastric, colorectal, renal, breast and cervical carcinomas, as well as non-Hodgkin, Hodgkin and nasal natural killer/T-cell lymphomas, although all three <i>TET</i> family genes are ubiquitously expressed in normal tissues. Ectopic expression of TET1 catalytic domain suppressed colony formation and induced apoptosis of tumor cells of multiple tissue types, supporting its role as a broad <i>bona fide</i> tumor suppressor. Furthermore, TET1 catalytic domain possessed demethylase activity in cancer cells, being able to inhibit the CpG methylation of tumor suppressor gene (TSG) promoters and reactivate their expression, such as <i>SLIT2, ZNF382</i> and <i>HOXA9</i>. As only infrequent mutations of <i>TET1</i> have been reported, compared to <i>TET2</i>, epigenetic silencing therefore appears to be the dominant mechanism for <i>TET1</i> inactivation in cancers, which also forms a feedback loop of CpG methylation during tumorigenesis.</p>',
'date' => '2016-05-26',
'pmid' => 'http://www.nature.com/articles/srep26591',
'doi' => '10.1038/srep26591',
'modified' => '2016-06-06 09:47:31',
'created' => '2016-06-06 09:47:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 68 => array(
'id' => '2836',
'name' => 'Biochemical reconstitution of TET1–TDG–BER-dependent active DNA demethylation reveals a highly coordinated mechanism',
'authors' => 'Weber AR, Krawczyk C, Robertson AB, Kuśnierczyk A, Vågbø CB, Schuermann D, Klungland A, Schär P',
'description' => '<p>Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten–eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET–TDG–BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.nature.com/ncomms/2016/160302/ncomms10806/full/ncomms10806.html',
'doi' => '10.1038/ncomms10806',
'modified' => '2016-03-08 10:25:46',
'created' => '2016-03-08 10:25:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 69 => array(
'id' => '2837',
'name' => 'Hydroxymethylation of microRNA-365-3p Regulates Nociceptive Behaviors via Kcnh2',
'authors' => 'Pan Z, Zhang M, Ma T, Xue Z-Y, Li G-F, Hao L-Y, Zhu L-J, Li Y-Q, Ding H-L, Cao J-L',
'description' => '<p id="p-3">DNA 5-hydroxylmethylcytosine (5hmC) catalyzed by ten-eleven translocation methylcytosine dioxygenase (TET) occurs abundantly in neurons of mammals. However, the <em>in vivo</em> causal link between TET dysregulation and nociceptive modulation has not been established. Here, we found that spinal TET1 and TET3 were significantly increased in the model of formalin-induced acute inflammatory pain, which was accompanied with the augment of genome-wide 5hmC content in spinal cord. Knockdown of spinal TET1 or TET3 alleviated the formalin-induced nociceptive behavior and overexpression of spinal TET1 or TET3 in naive mice produced pain-like behavior as evidenced by decreased thermal pain threshold. Furthermore, we found that TET1 or TET3 regulated the nociceptive behavior by targeting microRNA-365-3p (miR-365-3p). Formalin increased 5hmC in the miR-365-3p promoter, which was inhibited by knockdown of TET1 or TET3 and mimicked by overexpression of TET1 or TET3 in naive mice. Nociceptive behavior induced by formalin or overexpression of spinal TET1 or TET3 could be prevented by downregulation of miR-365-3p, and mimicked by overexpression of spinal miR-365-3p. Finally, we demonstrated that a potassium channel, voltage-gated eag-related subfamily H member 2 (<em>Kcnh2</em>), validated as a target of miR-365-3p, played a critical role in nociceptive modulation by spinal TET or miR-365-3p. Together, we concluded that TET-mediated hydroxymethylation of miR-365-3p regulates nociceptive behavior via <em>Kcnh2</em>.</p>
<p id="p-4"><strong>SIGNIFICANCE STATEMENT</strong> Mounting evidence indicates that epigenetic modifications in the nociceptive pathway contribute to pain processes and analgesia response. Here, we found that the increase of 5hmC content mediated by TET1 or TET3 in miR-365-3p promoter in the spinal cord is involved in nociceptive modulation through targeting a potassium channel, <em>Kcnh2</em>. Our study reveals a new epigenetic mechanism underlying nociceptive information processing, which may be a novel target for development of antinociceptive drugs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.jneurosci.org/content/36/9/2769.short',
'doi' => '10.1523/JNEUROSCI.3474-15.2016',
'modified' => '2016-03-08 10:40:48',
'created' => '2016-03-08 10:40:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 70 => array(
'id' => '3034',
'name' => 'Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism',
'authors' => 'Weber AR et al.',
'description' => '<p>Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten-eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET-TDG-BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.</p>',
'date' => '2016-03-02',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26932196',
'doi' => '',
'modified' => '2016-09-23 16:34:57',
'created' => '2016-09-23 16:34:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 71 => array(
'id' => '2833',
'name' => 'Genome-wide DNA methylation profile of developing deciduous tooth germ in miniature pigs',
'authors' => 'Su Y, Fan Z, Wu X, Li Y, Wang F, Zhang C, Wang J, Du J, Wang S',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND"><span class="highlight">DNA</span> <span class="highlight">methylation</span> is an important epigenetic modification critical to the regulation of gene expression during development. To date, little is known about the role of <span class="highlight">DNA</span> <span class="highlight">methylation</span> in <span class="highlight">tooth</span> development in large animal models. Thus, we carried out a comparative genomic analysis of <span class="highlight">genome-wide</span> <span class="highlight">DNA</span> <span class="highlight">methylation</span> profiles in E50 and E60 <span class="highlight">tooth</span> <span class="highlight">germ</span> from <span class="highlight">miniature</span> <span class="highlight">pigs</span> using methylated <span class="highlight">DNA</span> immunoprecipitation-sequencing (MeDIP-seq).</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We observed different <span class="highlight">DNA</span> <span class="highlight">methylation</span> patterns during the different developmental stages of pig <span class="highlight">tooth</span> <span class="highlight">germ</span>. A total of 2469 differentially methylated genes were identified. Functional analysis identified several signaling pathways and 104 genes that may be potential key regulators of pig <span class="highlight">tooth</span> development from E50 to E60.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">The present study provided a comprehensive analysis of the global <span class="highlight">DNA</span> <span class="highlight">methylation</span> pattern of <span class="highlight">tooth</span> <span class="highlight">germ</span> in <span class="highlight">miniature</span> <span class="highlight">pigs</span> and identified candidate genes that potentially regulate <span class="highlight">tooth</span> development from E50 to E60.</abstracttext></p>
</div>',
'date' => '2016-02-24',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26911717',
'doi' => '10.1186/s12864-016-2485-9',
'modified' => '2016-03-01 10:35:04',
'created' => '2016-03-01 10:35:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 72 => array(
'id' => '2834',
'name' => 'Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.',
'authors' => 'Rebollo R, Mager DL',
'description' => '<p>Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method used in our laboratory to detect, quantify, and compare mammalian endogenous retrovirus DNA methylation. More specifically we describe methylated DNA immunoprecipitation (MeDIP) followed by quantitative PCR.</p>',
'date' => '2016-02-20',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26895065',
'doi' => '10.1007/978-1-4939-3372-3_23',
'modified' => '2016-03-02 10:37:15',
'created' => '2016-03-02 10:22:16',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 73 => array(
'id' => '2815',
'name' => 'The dual specificity phosphatase 2 gene is hypermethylated in human cancer and regulated by epigenetic mechanisms',
'authors' => 'Tanja Haag, Antje M. Richter, Martin B. Schneider, Adriana P. Jiménez and Reinhard H. Dammann',
'description' => '<p><span>Dual specificity phosphatases are a class of tumor-associated proteins involved in the negative regulation of the MAP kinase pathway. Downregulation of the </span><em xmlns="" class="EmphasisTypeItalic">dual specificity phosphatase 2</em><span> (</span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span>) has been reported in cancer. Epigenetic silencing of tumor suppressor genes by abnormal promoter methylation is a frequent mechanism in oncogenesis. It has been shown that the epigenetic factor CTCF is involved in the regulation of tumor suppressor genes.</span></p>
<p><span>We analyzed the promoter hypermethylation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> in human cancer, including primary Merkel cell carcinoma by bisulfite restriction analysis and pyrosequencing. Moreover we analyzed the impact of a DNA methyltransferase inhibitor (5-Aza-dC) and CTCF on the epigenetic regulation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> by qRT-PCR, promoter assay, chromatin immuno-precipitation and methylation analysis.</span></span></p>
<p><span><span>Here we report a significant tumor-specific hypermethylation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> in primary Merkel cell carcinoma (</span><em xmlns="" class="EmphasisTypeItalic">p</em><span> = 0.05). An increase in methylation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> was also found in 17 out of 24 (71 %) cancer cell lines, including skin and lung cancer. Treatment of cancer cells with 5-Aza-dC induced </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression by its promoter demethylation, Additionally we observed that CTCF induces </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression in cell lines that exhibit silencing of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span>. This reactivation was accompanied by increased CTCF binding and demethylation of the </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> promoter.</span></span></span></p>
<p><span><span><span>Our data show that aberrant epigenetic inactivation of <em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> occurs in carcinogenesis and that CTCF is involved in the epigenetic regulation of </span><em xmlns="" class="EmphasisTypeItalic">DUSP2</em><span> expression.</span></span></span></span></p>',
'date' => '2016-02-01',
'pmid' => 'http://pubmed.gov/26833217',
'doi' => '10.1186/s12885-016-2087-6',
'modified' => '2016-03-09 16:34:51',
'created' => '2016-02-09 14:47:36',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 74 => array(
'id' => '2859',
'name' => 'Role of Growth Arrest and DNA Damage-Inducible, Beta in Alcohol-Drinking Behaviors',
'authors' => 'Gavin DP, Kusumo H, Zhang H, Guidotti A, Pandey SC',
'description' => '<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">The contribution of epigenetic factors, such as histone acetylation and DNA methylation, to the regulation of alcohol-drinking behavior has been increasingly recognized over the last several years. GADD45b is a protein demonstrated to be involved in DNA demethylation at neurotrophic factor gene promoters, including at brain-derived neurotrophic factor (Bdnf) which has been highly implicated in alcohol-drinking behavior.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">DNA methyltransferase-1 (Dnmt1), 3a, and 3b, and Gadd45a, b, and g mRNA were measured in the nucleus accumbens (NAc) and ventral tegmental areas of high ethanol (EtOH) consuming C57BL/6J (C57) and low alcohol consuming DBA/2J (DBA) mice using quantitative reverse transcriptase polymerase chain reaction (PCR). In the NAc, GADD45b protein was measured via immunohistochemistry and Bdnf9a mRNA using in situ PCR. Bdnf9a promoter histone H3 acetylated at lysines 9 and 14 (H3K9,K14ac) was measured using chromatin immunoprecipitation, and 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) using methylated DNA immunoprecipitation. Alcohol-drinking behavior was evaluated in Gadd45b haplodeficient (+/-) and null mice (-/-) utilizing drinking-in-the-dark (DID) and 2-bottle free-choice paradigms.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">C57 mice had lower levels of Gadd45b and g mRNA and GADD45b protein in the NAc relative to the DBA strain. C57 mice had lower NAc shell Bdnf9a mRNA levels, Bdnf9a promoter H3K9,K14ac, and higher Bdnf9a promoter 5HMC and 5MC. Acute EtOH increased GADD45b protein, Bdnf9a mRNA, and histone acetylation and decreased 5HMC in C57 mice. Gadd45b +/- mice displayed higher drinking behavior relative to wild-type littermates in both DID and 2-bottle free-choice paradigms.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">These data indicate the importance of the DNA demethylation pathway and its interactions with histone posttranslational modifications in alcohol-drinking behavior. Further, we suggest that lower DNA demethylation protein GADD45b levels may affect Bdnf expression possibly leading to altered alcohol-drinking behavior.</abstracttext></p>',
'date' => '2016-02-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26842245',
'doi' => ' 10.1111/acer.12965',
'modified' => '2016-03-15 16:37:22',
'created' => '2016-03-15 16:37:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 75 => array(
'id' => '2975',
'name' => 'Protocol for Methylated DNA Immunoprecipitation (MeDIP) Analysis',
'authors' => 'Karpova NN et al.',
'description' => '<p>DNA methylation is a fundamental epigenetic mechanism for silencing gene expression by either modifying chromatin structure to a repressive state or interfering with the transcription factors’ binding. DNA methylation primarily occurs at the position C5 of a cytosine ring mainly in the context of CpG dinucleotides. The modification can be recognized both in vivo and in vitro by the methyl-CpG binding proteins (MBPs) as well as in vitro by an antibody raised against 5-methylcytosine (5mC). This chapter describes different MBPs and introduces a standard methylated DNA immunoprecipitation (MeDIP) method, which is based on using the anti-5mC antibody to isolate methylated DNA fragments for subsequent locus-specific DNA methylation analysis. The MeDIP-generated DNA can be used as well for methylation profiling on a genome scale using array-based (MeDIP-chip) and high-throughput (MeDIP-seq) technologies.</p>',
'date' => '2016-02-01',
'pmid' => 'http://link.springer.com/protocol/10.1007/978-1-4939-2754-8_6',
'doi' => '10.1007/978-1-4939-2754-8_6',
'modified' => '2016-07-07 09:35:44',
'created' => '2016-07-07 09:35:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 76 => array(
'id' => '2844',
'name' => 'De novo DNA methylation drives 5hmC accumulation in mouse zygotes',
'authors' => 'Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, Koseki H, Sasaki H, Hajkova P',
'description' => '<p>Zygotic epigenetic reprogramming entails genome-wide DNA demethylation that is accompanied by Tet methylcytosine dioxygenase 3 (Tet3)-driven oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC; refs ,,,). Here we demonstrate using detailed immunofluorescence analysis and ultrasensitive LC-MS-based quantitative measurements that the initial loss of paternal 5mC does not require 5hmC formation. Small-molecule inhibition of Tet3 activity, as well as genetic ablation, impedes 5hmC accumulation in zygotes without affecting the early loss of paternal 5mC. Instead, 5hmC accumulation is dependent on the activity of zygotic Dnmt3a and Dnmt1, documenting a role for Tet3-driven hydroxylation in targeting de novo methylation activities present in the early embryo. Our data thus provide further insights into the dynamics of zygotic reprogramming, revealing an intricate interplay between DNA demethylation, de novo methylation and Tet3-driven hydroxylation.</p>',
'date' => '2016-01-11',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26751286',
'doi' => '10.1038/ncb3296',
'modified' => '2016-03-09 17:32:33',
'created' => '2016-03-09 17:32:33',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 77 => array(
'id' => '2860',
'name' => 'DNA methylation profiling: comparison of genome-wide sequencing methods and the Infinium Human Methylation 450 Bead Chip',
'authors' => 'Walker DL, Bhagwate AV, Baheti S, Smalley RL, Hilker CA, Sun Z, Cunningham JM',
'description' => '<div class="">
<h4>AIMS:</h4>
<p><abstracttext label="AIMS" nlmcategory="OBJECTIVE">To compare the performance of four sequence-based and one microarray methods for DNA methylation profiling.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">DNA from two cell lines were profiled by reduced representation bisulfite sequencing, methyl capture sequencing (SS-Meth Seq), NimbleGen SeqCapEpi CpGiant(Nimblegen MethSeq), methylated DNA immunoprecipitation (MeDIP) and the Human Methylation 450 Bead Chip (Meth450K).</abstracttext></p>
<h4>RESULTS & CONCLUSION:</h4>
<p><abstracttext label="RESULTS & CONCLUSION" nlmcategory="CONCLUSIONS">Despite differences in genome-wide coverage, high correlation and concordance were observed between different methods. Significant overlap of differentially methylated regions was identified between sequenced-based platforms. MeDIP provided the best coverage for the whole genome and gene body regions, while RRBS and Nimblegen MethSeq were superior for CpGs in CpG islands and promoters. Methylation analyses can be achieved by any of the five methods but understanding their differences may better address the research question being posed.</abstracttext></p>
</div>',
'date' => '2015-12-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26192535',
'doi' => '10.2217/EPI.15.64',
'modified' => '2016-03-16 11:06:05',
'created' => '2016-03-16 11:06:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 78 => array(
'id' => '2931',
'name' => 'Oxidative DNA damage in mouse sperm chromosomes: Size matters.',
'authors' => 'Kocer A et al.',
'description' => '<p>Normal embryo and foetal development as well as the health of the progeny are mostly dependent on gamete nuclear integrity. In the present study, in order to characterize more precisely oxidative DNA damage in mouse sperm we used two mouse models that display high levels of sperm oxidative DNA damage, a common alteration encountered both in in vivo and in vitro reproduction. Immunoprecipitation of oxidized sperm DNA coupled to deep sequencing showed that mouse chromosomes may be largely affected by oxidative alterations. We show that the vulnerability of chromosomes to oxidative attack inversely correlated with their size and was not linked to their GC richness. It was neither correlated with the chromosome content in persisting nucleosomes nor associated with methylated sequences. A strong correlation was found between oxidized sequences and sequences rich in short interspersed repeat elements (SINEs). Chromosome position in the sperm nucleus as revealed by fluorescent in situ hybridization appears to be a confounder. These data map for the first time fragile mouse sperm chromosomal regions when facing oxidative damage that may challenge the repair mechanisms of the oocyte post-fertilization.</p>',
'date' => '2015-12-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26510519',
'doi' => '10.1016/j.freeradbiomed.2015.10.419',
'modified' => '2016-05-19 10:18:24',
'created' => '2016-05-19 10:18:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 79 => array(
'id' => '2944',
'name' => 'Immunohistochemical Detection of Oxidized Forms of 5-Methylcytosine in Embryonic and Adult Brain Tissue',
'authors' => 'Abakir A et al.',
'description' => '<p>DNA methylation (5-methylcytosine, 5mC) is a major epigenetic modification of the eukaryotic genome associated with gene repression. Ten-eleven translocation proteins (Tet1/2/3) can oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Recent studies demonstrate that 5hmC is particularly enriched in neuronal cells and imply the involvement of this mark in transcriptional regulation taking place within the mammalian brain. Although a number of biochemical and antibody-based approaches have been successfully used to study the global content and genomic distributions of 5hmC in various contexts, most of these techniques do not provide any spatial information on the levels of this mark in different cell types. Here we describe a method of sensitive immunochemical detection of 5hmC/5fC/5caC in brain tissue based on the use of peroxidase-conjugated secondary antibodies and tyramide signal amplification. This technique can be instrumental in determining the relative enrichments of oxidized forms of 5mC in different brain cell types, effectively complementing other established approaches to investigate the functions of these marks in embryonic and adult brain.</p>',
'date' => '2015-09-02',
'pmid' => 'http://link.springer.com/protocol/10.1007%2F978-1-4939-2754-8_8',
'doi' => ' Print ISBN 978-1-4939-2753-1 Online ISBN 978-1-4939-2754-8 Series Title Neuromethods Series Volume 105 Series ISSN 0893-2336 Publisher Springer New York Copyright Holder Springer Science+Business Media New York Additional Lin',
'modified' => '2016-06-08 10:16:17',
'created' => '2016-06-08 10:16:17',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 80 => array(
'id' => '2847',
'name' => 'Gadd45b and N-methyl-D-aspartate induced DNA demethylation in postmitotic neurons.',
'authors' => 'Gavin DP, Kusumo H, Sharma RP, Guizzetti M, Guidotti A, Pandey SC.',
'description' => '<p><strong>AIM:</strong> In nondividing neurons examine the role of Gadd45b in active 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) removal at a gene promoter highly implicated in mental illnesses and cognition, Bdnf.</p>
<p><strong>MATERIALS & METHODS:</strong> Mouse primary cortical neuronal cultures with and without Gadd45b siRNA transfection were treated with N-methyl-d-aspartate (NMDA). Expression changes of genes reportedly involved in DNA demethylation, Bdnf mRNA and protein and 5MC and 5HMC at Bdnf promoters were measured.</p>
<p><strong>RESULTS:</strong> Gadd45b siRNA transfection in neurons abolishes the NMDA-induced increase in Bdnf IXa mRNA and reductions in 5MC and 5HMC at the Bdnf IXa promoter.</p>
<p><strong>CONCLUSION:</strong> These results contribute to our understanding of DNA demethylation mechanisms in neurons, and its role in regulating NMDA responsive genes implicated in mental illnesses.</p>',
'date' => '2015-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/26111030',
'doi' => '10.2217/epi.15.12',
'modified' => '2016-03-11 16:02:08',
'created' => '2016-03-11 15:47:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 81 => array(
'id' => '2879',
'name' => 'Active human nucleolar organizer regions are interspersed with inactive rDNA repeats in normal and tumor cells.',
'authors' => 'Zillner K, Komatsu J, Filarsky K, Kalepu R, Bensimon A, Németh A',
'description' => '<div class="">
<h4>AIM:</h4>
<p><abstracttext label="AIM" nlmcategory="OBJECTIVE">The synthesis of rRNA is a key determinant of normal and malignant cell growth and subject to epigenetic regulation. Yet, the epigenomic features of rDNA arrays clustered in nucleolar organizer regions are largely unknown. We set out to explore for the first time how DNA methylation is distributed on individual rDNA arrays.</abstracttext></p>
<h4>MATERIALS & METHODS:</h4>
<p><abstracttext label="MATERIALS & METHODS" nlmcategory="METHODS">Here we combined immunofluorescence detection of DNA modifications with fluorescence hybridization of single DNA fibers, metaphase immuno-FISH and methylation-sensitive restriction enzyme digestions followed by Southern blot.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We found clustering of both hypomethylated and hypermethylated repeat units and hypermethylation of noncanonical rDNA in IMR90 fibroblasts and HCT116 colorectal carcinoma cells. Surprisingly, we also found transitions between hypo- and hypermethylated rDNA repeat clusters on single DNA fibers.</abstracttext></p>
<h4>CONCLUSION:</h4>
<p><abstracttext label="CONCLUSION" nlmcategory="CONCLUSIONS">Collectively, our analyses revealed co-existence of different epialleles on individual nucleolar organizer regions and showed that epi-combing is a valuable approach to analyze epigenomic patterns of repetitive DNA.</abstracttext></p>
</div>',
'date' => '2015-06-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26077426',
'doi' => '10.2217/epi.14.93',
'modified' => '2016-04-05 09:44:29',
'created' => '2016-04-05 09:44:29',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 82 => array(
'id' => '2790',
'name' => 'Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency.',
'authors' => 'Chen H, Aksoy I, Gonnot F, Osteil P, Aubry M, Hamela C, Rognard C, Hochard A, Voisin S, Fontaine E, Mure M, Afanassieff M, Cleroux E, Guibert S, Chen J, Vallot C, Acloque H, Genthon C, Donnadieu C, De Vos J, Sanlaville D, Guérin JF, Weber M, Stanton LW, R',
'description' => 'Leukemia inhibitory factor (LIF)/STAT3 signalling is a hallmark of naive pluripotency in rodent pluripotent stem cells (PSCs), whereas fibroblast growth factor (FGF)-2 and activin/nodal signalling is required to sustain self-renewal of human PSCs in a condition referred to as the primed state. It is unknown why LIF/STAT3 signalling alone fails to sustain pluripotency in human PSCs. Here we show that the forced expression of the hormone-dependent STAT3-ER (ER, ligand-binding domain of the human oestrogen receptor) in combination with 2i/LIF and tamoxifen allows human PSCs to escape from the primed state and enter a state characterized by the activation of STAT3 target genes and long-term self-renewal in FGF2- and feeder-free conditions. These cells acquire growth properties, a gene expression profile and an epigenetic landscape closer to those described in mouse naive PSCs. Together, these results show that temporarily increasing STAT3 activity is sufficient to reprogramme human PSCs to naive-like pluripotent cells.',
'date' => '2015-05-13',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25968054',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 83 => array(
'id' => '2678',
'name' => 'Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells.',
'authors' => 'Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, Pop R, Reyon D, Tsai SQ, Mallard W, Joung JK, Rinn JL, Gnirke A, Meissner A',
'description' => 'DNA methylation is a key epigenetic modification involved in regulating gene expression and maintaining genomic integrity. Here we inactivated all three catalytically active DNA methyltransferases (DNMTs) in human embryonic stem cells (ESCs) using CRISPR/Cas9 genome editing to further investigate the roles and genomic targets of these enzymes. Disruption of DNMT3A or DNMT3B individually as well as of both enzymes in tandem results in viable, pluripotent cell lines with distinct effects on the DNA methylation landscape, as assessed by whole-genome bisulfite sequencing. Surprisingly, in contrast to findings in mouse, deletion of DNMT1 resulted in rapid cell death in human ESCs. To overcome this immediate lethality, we generated a doxycycline-responsive tTA-DNMT1* rescue line and readily obtained homozygous DNMT1-mutant lines. However, doxycycline-mediated repression of exogenous DNMT1* initiates rapid, global loss of DNA methylation, followed by extensive cell death. Our data provide a comprehensive characterization of DNMT-mutant ESCs, including single-base genome-wide maps of the targets of these enzymes.',
'date' => '2015-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25822089',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 84 => array(
'id' => '2477',
'name' => 'Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes.',
'authors' => 'Li L, Zhang Y, Fan Y, Sun K, Su X, Du Z, Tsao SW, Loh TK, Sun H, Chan AT, Zeng YX, Chan WY, Chan FK, Tao Q',
'description' => 'Aims: Nasopharyngeal carcinoma (NPC) is a common tumor consistently associated with Epstein-Barr virus infection and prevalent in South China, including Hong Kong, and southeast Asia. Current genomic sequencing studies found only rare mutations in NPC, indicating its critical epigenetic etiology, while no epigenome exists for NPC as yet. Materials & methods: We profiled the methylomes of NPC cell lines and primary tumors, together with normal nasopharyngeal epithelial cells, using methylated DNA immunoprecipitation (MeDIP). Results: We observed extensive, genome-wide methylation of cellular genes. Epigenetic disruption of Wnt, MAPK, TGF-β and Hedgehog signaling pathways was detected. Methylation of Wnt signaling regulators (SFRP1, 2, 4 and 5, DACT2, DKK2 and DKK3) was frequently detected in tumor and nasal swab samples from NPC patients. Functional studies showed that these genes are bona fide tumor-suppressor genes for NPC. Conclusion: The NPC methylome shows a special high-degree CpG methylation epigenotype, similar to the Epstein-Barr virus-infected gastric cancer, indicating a critical epigenetic etiology for NPC pathogenesis.',
'date' => '2015-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25479246',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 85 => array(
'id' => '2613',
'name' => 'CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.',
'authors' => 'Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC',
'description' => 'BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the underlying epigenetic signatures involved in its regulation. RESULTS: NOS3 and STAT3 mRNA levels were elevated in HUAEC of patients who suffered from placental insufficiency. 5-hydroxymethylcytosine, H3K9ac and Histone 2A (H2A).Zac at the NOS3 transcription start site directly correlated with NOS3 mRNA levels. Concomitantly, we observed entangled histone acetylation patterns and NOS3 response upon hypoxic conditions in vitro. Knock-down of either NOS3 or STAT3 by RNAi provided evidence for a functional NOS3/STAT3 relationship. Moreover, we recognized massive turnover of Stat3 at a discrete binding site in the NOS3 promoter. Interestingly, induced hyperacetylation resulted in short-termed increase of NOS3 mRNA followed by deferred decrease indicating that NOS3 expression could become self-attenuated by co-expressed intronic 27 nt-ncRNA. Reporter assay results and phylogenetic analyses enabled us to propose a novel model for STAT3-3'-UTR targeting by this 27-nt-ncRNA. CONCLUSIONS: An adverse intrauterine environment leads to adaptive changes of NOS3 expression. Apparently, a rapid NOS3 self-limiting response upon ectopic triggers co-exists with longer termed expression changes in response to placental insufficiency involving differential epigenetic signatures. Their persistence might contribute to impaired vascular endothelial response and consequently increase the risk of cardiovascular disease later in life.',
'date' => '2015-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25699114',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 86 => array(
'id' => '2437',
'name' => 'Acute Depletion Redefines the Division of Labor among DNA Methyltransferases in Methylating the Human Genome.',
'authors' => 'Tiedemann RL, Putiri EL, Lee JH, Hlady RA, Kashiwagi K, Ordog T, Zhang Z, Liu C, Choi JH, Robertson KD',
'description' => 'Global patterns of DNA methylation, mediated by the DNA methyltransferases (DNMTs), are disrupted in all cancers by mechanisms that remain largely unknown, hampering their development as therapeutic targets. Combinatorial acute depletion of all DNMTs in a pluripotent human tumor cell line, followed by epigenome and transcriptome analysis, revealed DNMT functions in fine detail. DNMT3B occupancy regulates methylation during differentiation, whereas an unexpected interplay was discovered in which DNMT1 and DNMT3B antithetically regulate methylation and hydroxymethylation in gene bodies, a finding confirmed in other cell types. DNMT3B mediated non-CpG methylation, whereas DNMT3L influenced the activity of DNMT3B toward non-CpG versus CpG site methylation. Altogether, these data reveal functional targets of each DNMT, suggesting that isoform selective inhibition would be therapeutically advantageous.',
'date' => '2014-11-20',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25453758',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 87 => array(
'id' => '2368',
'name' => 'A B-cell targeting virus disrupts potentially protective genomic methylation patterns in lymphoid tissue by increasing global 5-hydroxmethylcytosine levels',
'authors' => 'Ciccone NA, Mwangi W, Ruzov A, Smith LP, Butter C, Nair V',
'description' => 'The mechanisms by which viruses modulate the immune system include changes in host genomic methylation. 5-hydroxmethylcytosine (5hmC) is the catalytic product of the Tet (Ten-11 translocation) family of enzymes and may serve as an intermediate of DNA demethylation. Recent reports suggest that 5hmC may confer consequences on cellular events including the pathogenesis of disease; in order to explore this possibility further we investigated both 5-methylcytosine (5mC) and 5hmC levels in healthy and diseased chicken bursas of Fabricius. We discovered that embryonic B-cells have high 5mC content while 5hmC decreases during bursa development. We propose that a high 5mC level protects from the mutagenic activity of the B-cell antibody diversifying enzyme activation induced deaminase (AID). In support of this view, AID mRNA increases significantly within the developing bursa from embryonic to post hatch stages while mRNAs that encode Tet family members 1 and 2 reduce over the same period. Moreover, our data revealed that infectious bursal disease virus (IBDV) disrupts this genomic methylation pattern causing a global increase in 5hmC levels in a mechanism that may involve increased Tet 1 and 2 mRNAs. To our knowledge this is the first time that a viral infection has been observed to cause global increases in genomic 5hmC within infected host tissues, underlining a mechanism that may involve the induction of B-cell genomic instability and cell death to facilitate viral egress.',
'date' => '2014-10-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/25338704',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 88 => array(
'id' => '2350',
'name' => 'Spontaneous sleep-wake cycle and sleep deprivation differently induce Bdnf1, Bdnf4 and Bdnf9a DNA methylation and transcripts levels in the basal forebrain and frontal cortex in rats.',
'authors' => 'Ventskovska O, Porkka-Heiskanen T, Karpova NN',
'description' => 'Brain-derived neurotrophic factor (Bdnf) regulates neuronal plasticity, slow wave activity and sleep homeostasis. Environmental stimuli control Bdnf expression through epigenetic mechanisms, but there are no data on epigenetic regulation of Bdnf by sleep or sleep deprivation. Here we investigated whether 5-methylcytosine (5mC) DNA modification at Bdnf promoters p1, p4 and p9 influences Bdnf1, Bdnf4 and Bdnf9a expression during the normal inactive phase or after sleep deprivation (SD) (3, 6 and 12 h, end-times being ZT3, ZT6 and ZT12) in rats in two brain areas involved in sleep regulation, the basal forebrain and cortex. We found a daytime variation in cortical Bdnf expression: Bdnf1 expression was highest at ZT6 and Bdnf4 lowest at ZT12. Such variation was not observed in the basal forebrain. Also Bdnf p1 and p9 methylation levels differed only in the cortex, while Bdnf p4 methylation did not vary in either area. Factorial analysis revealed that sleep deprivation significantly induced Bdnf1 and Bdnf4 with the similar pattern for Bdnf9a in both basal forebrain and cortex; 12 h of sleep deprivation decreased 5mC levels at the cortical Bdnf p4 and p9. Regression analysis between the 5mC promoter levels and the corresponding Bdnf transcript expression revealed significant negative correlations for the basal forebrain Bdnf1 and cortical Bdnf9a transcripts in only non-deprived rats, while these correlations were lost after sleep deprivation. Our results suggest that Bdnf transcription during the light phase of undisturbed sleep-wake cycle but not after SD is regulated at least partially by brain site-specific DNA methylation.',
'date' => '2014-09-15',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25223586',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 89 => array(
'id' => '2109',
'name' => 'Transient accumulation of 5-carboxylcytosine indicates involvement of active demethylation in lineage specification of neural stem cells.',
'authors' => 'Wheldon LM, Abakir A, Ferjentsik Z, Dudnakova T, Strohbuecker S, Christie D, Dai N, Guan S, Foster JM, Corrêa IR, Loose M, Dixon JE, Sottile V, Johnson AD, Ruzov A',
'description' => '5-Methylcytosine (5mC) is an epigenetic modification involved in regulation of gene activity during differentiation. Tet dioxygenases oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). Both 5fC and 5caC can be excised from DNA by thymine-DNA glycosylase (TDG) followed by regeneration of unmodified cytosine via the base excision repair pathway. Despite evidence that this mechanism is operative in embryonic stem cells, the role of TDG-dependent demethylation in differentiation and development is currently unclear. Here, we demonstrate that widespread oxidation of 5hmC to 5caC occurs in postimplantation mouse embryos. We show that 5fC and 5caC are transiently accumulated during lineage specification of neural stem cells (NSCs) in culture and in vivo. Moreover, 5caC is enriched at the cell-type-specific promoters during differentiation of NSCs, and TDG knockdown leads to increased 5fC/5caC levels in differentiating NSCs. Our data suggest that active demethylation contributes to epigenetic reprogramming determining lineage specification in embryonic brain.',
'date' => '2014-06-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24882006',
'doi' => '',
'modified' => '2015-07-24 15:39:03',
'created' => '2015-07-24 15:39:03',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 90 => array(
'id' => '2448',
'name' => 'Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring.',
'authors' => 'Itzhak Y, Ergui I, Young JI',
'description' => 'The high rate of methamphetamine (METH) abuse among young adults and women of childbearing age makes it imperative to determine the long-term effects of METH exposure on the offspring. We hypothesized that parental METH exposure modulates offspring behavior by disrupting epigenetic programming of gene expression in the brain. To simulate the human pattern of drug use, male and female C57Bl/6J mice were exposed to escalating doses of METH or saline from adolescence through adulthood; following mating, females continue to receive drug or saline through gestational day 17. F1 METH male offspring showed enhanced response to cocaine-conditioned reward and hyperlocomotion. Both F1 METH male and female offspring had reduced response to conditioned fear. Cross-fostering experiments have shown that certain behavioral phenotypes were modulated by maternal care of either METH or saline dams. Analysis of offspring hippocampal DNA methylation showed differentially methylated regions as a result of both METH in utero exposure and maternal care. Our results suggest that behavioral phenotypes and epigenotypes of offspring that were exposed to METH in utero are vulnerable to (a) METH exposure during embryonic development, a period when wide epigenetic reprogramming occurs, and (b) postnatal maternal care.Molecular Psychiatry advance online publication, 18 February 2014; doi:10.1038/mp.2014.7.',
'date' => '2014-02-18',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24535458',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 91 => array(
'id' => '1843',
'name' => 'Alterations of epigenetic signatures in hepatocyte nuclear factor 4α deficient mouse liver determined by improved ChIP-qPCR and (h)MeDIP-qPCR assays.',
'authors' => 'Zhang Q, Lei X, Lu H',
'description' => 'Hepatocyte nuclear factor 4α (HNF4α) is a liver-enriched transcription factor essential for liver development and function. In hepatocytes, HNF4α regulates a large number of genes important for nutrient/xenobiotic metabolism and cell differentiation and proliferation. Currently, little is known about the epigenetic mechanism of gene regulation by HNF4α. In this study, the global and specific alterations at the selected gene loci of representative histone modifications and DNA methylations were investigated in Hnf4a-deficient female mouse livers using the improved MeDIP-, hMeDIP- and ChIP-qPCR assay. Hnf4a deficiency significantly increased hepatic total IPed DNA fragments for histone H3 lysine-4 dimethylation (H3K4me2), H3K4me3, H3K9me2, H3K27me3 and H3K4 acetylation, but not for H3K9me3, 5-methylcytosine,or 5-hydroxymethylcytosine. At specific gene loci, the relative enrichments of histone and DNA modifications were changed to different degree in Hnf4a-deficient mouse liver. Among the epigenetic signatures investigated, changes in H3K4me3 correlated the best with mRNA expression. Additionally, Hnf4a-deficient livers had increased mRNA expression of histone H1.2 and H3.3 as well as epigenetic modifiers Dnmt1, Tet3, Setd7, Kmt2c, Ehmt2, and Ezh2. In conclusion, the present study provides convenient improved (h)MeDIP- and ChIP-qPCR assays for epigenetic study. Hnf4a deficiency in young-adult mouse liver markedly alters histone methylation and acetylation, with fewer effects on DNA methylation and 5-hydroxymethylation. The underlying mechanism may be the induction of epigenetic enzymes responsible for the addition/removal of the epigenetic signatures, and/or the loss of HNF4α per se as a key coordinator for epigenetic modifiers.',
'date' => '2014-01-10',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24427299',
'doi' => '',
'modified' => '2015-07-24 15:39:01',
'created' => '2015-07-24 15:39:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 92 => array(
'id' => '1773',
'name' => 'Peroxisome proliferator-activated receptor γ regulates genes involved in insulin/insulin-like growth factor signaling and lipid metabolism during adipogenesis through functionally distinct enhancer classes.',
'authors' => 'Oger F, Dubois-Chevalier J, Gheeraert C, Avner S, Durand E, Froguel P, Salbert G, Staels B, Lefebvre P, Eeckhoute J',
'description' => 'The nuclear receptor peroxisome proliferator-activated receptor (PPAR) is a transcription factor whose expression is induced during adipogenesis and that is required for the acquisition and control of mature adipocyte functions. Indeed, PPAR induces the expression of genes involved in lipid synthesis and storage through enhancers activated during adipocyte differentiation. Here, we show that PPAR also binds to enhancers already active in preadipocytes as evidenced by an active chromatin state including lower DNA methylation levels despite higher CpG content. These constitutive enhancers are linked to genes involved in the insulin/insulin-like growth factor signaling pathway that are transcriptionally induced during adipogenesis but to a lower extent than lipid metabolism genes, because of stronger basal expression levels in preadipocytes. This is consistent with the sequential involvement of hormonal sensitivity and lipid handling during adipocyte maturation and correlates with the chromatin structure dynamics at constitutive and activated enhancers. Interestingly, constitutive enhancers are evolutionary conserved and can be activated in other tissues, in contrast to enhancers controlling lipid handling genes whose activation is more restricted to adipocytes. Thus, PPAR utilizes both broadly active and cell type-specific enhancers to modulate the dynamic range of activation of genes involved in the adipogenic process.',
'date' => '2014-01-10',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24288131',
'doi' => '',
'modified' => '2015-07-24 15:39:01',
'created' => '2015-07-24 15:39:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 93 => array(
'id' => '1572',
'name' => 'Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model.',
'authors' => 'Yafune A, Kawai M, Itahashi M, Kimura M, Nakane F, Mitsumori K, Shibutani M',
'description' => 'Disruptive epigenetic gene control has been shown to be involved in carcinogenesis. To identify key molecules in piperonyl butoxide (PBO)-induced hepatocarcinogenesis, we searched hypermethylated genes using CpG island (CGI) microarrays in non-neoplastic liver cells as a source of proliferative lesions at 25 weeks after tumor promotion with PBO using mice. We further performed methylation-specific polymerase chain reaction (PCR), real-time reverse transcription PCR, and immunohistochemical analysis in PBO-promoted liver tissues. Ebp4.1, Wdr6 and Cmtm6 increased methylation levels in the promoter region by PBO promotion, although Cmtm6 levels were statistically non-significant. These results suggest that PBO promotion may cause altered epigenetic gene regulation in non-neoplastic liver cells surrounding proliferative lesions to allow the facilitation of hepatocarcinogenesis. Both Wdr6 and Cmtm6 showed decreased expression in non-neoplastic liver cells in contrast to positive immunoreactivity in the majority of proliferative lesions produced by PBO promotion. These results suggest that both Wdr6 and Cmtm6 were spared from epigenetic gene modification in proliferative lesions by PBO promotion in contrast to the hypermethylation-mediated downregulation in surrounding liver cells. Considering the effective detection of proliferative lesions, these molecules could be used as detection markers of hepatocellular proliferative lesions and played an important role in hepatocarcinogenesis.',
'date' => '2013-10-09',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23968726',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 94 => array(
'id' => '1569',
'name' => 'Genome-wide screening identifies Plasmodium chabaudi-induced modifications of DNA methylation status of Tlr1 and Tlr6 gene promoters in liver, but not spleen, of female C57BL/6 mice.',
'authors' => 'Al-Quraishy S, Dkhil MA, Abdel-Baki AA, Delic D, Santourlidis S, Wunderlich F',
'description' => '<p>Epigenetic reprogramming of host genes via DNA methylation is increasingly recognized as critical for the outcome of diverse infectious diseases, but information for malaria is not yet available. Here, we investigate the effect of blood-stage malaria of Plasmodium chabaudi on the DNA methylation status of host gene promoters on a genome-wide scale using methylated DNA immunoprecipitation and Nimblegen microarrays containing 2,000 bp oligonucleotide features that were split into -1,500 to -500 bp Ups promoters and -500 to +500 bp Cor promoters, relative to the transcription site, for evaluation of differential DNA methylation. Gene expression was analyzed by Agilent and Affymetrix microarray technology. Challenging of female C57BL/6 mice with 10(6) P. chabaudi-infected erythrocytes resulted in a self-healing outcome of infections with peak parasitemia on day 8 p.i. These infections induced organ-specific modifications of DNA methylation of gene promoters. Among the 17,354 features on Nimblegen arrays, only seven gene promoters were identified to be hypermethylated in the spleen, whereas the liver exhibited 109 hyper- and 67 hypomethylated promoters at peak parasitemia in comparison with non-infected mice. Among the identified genes with differentially methylated Cor-promoters, only the 7 genes Pigr, Ncf1, Klkb1, Emr1, Ndufb11, and Tlr6 in the liver and Apol6 in the spleen were detected to have significantly changed their expression. Remarkably, the Cor promoter of the toll-like receptor Tlr6 became hypomethylated and Tlr6 expression increased by 3.4-fold during infection. Concomitantly, the Ups promoter of the Tlr1 was hypermethylated, but Tlr1 expression also increased by 11.3-fold. TLR6 and TLR1 are known as auxillary receptors to form heterodimers with TLR2 in plasma membranes of macrophages, which recognize different pathogen-associated molecular patterns (PAMPs), as, e.g., intact 3-acyl and sn-2-lyso-acyl glycosylphosphatidylinositols of P. falciparum, respectively. Our data suggest therefore that malaria-induced epigenetic fine-tuning of Tlr6 and Tlr1 through DNA methylation of their gene promoters in the liver is critically important for initial recognition of PAMPs and, thus, for the final self-healing outcome of blood-stage infections with P. chabaudi malaria.</p>',
'date' => '2013-08-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23949311',
'doi' => '',
'modified' => '2017-10-10 10:37:58',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 95 => array(
'id' => '1466',
'name' => 'Characterization of the DNA methylome and its interindividual variation in human peripheral blood monocytes.',
'authors' => 'Shen H, Qiu C, Li J, Tian Q, Deng HW',
'description' => 'AIM: Peripheral blood monocytes (PBMs) play multiple and critical roles in the immune response, and abnormalities in PBMs have been linked to a variety of human disorders. However, the DNA methylation landscape in PBMs is largely unknown. In this study, we characterized epigenome-wide DNA methylation profiles in purified PBMs. MATERIALS & METHODS: PBMs were isolated from freshly collected peripheral blood from 18 unrelated healthy postmenopausal Caucasian females. Epigenome-wide DNA methylation profiles (the methylome) were characterized by using methylated DNA immunoprecipitation combined with high-throughput sequencing. RESULTS: Distinct patterns were revealed at different genomic features. For instance, promoters were commonly (∼58%) found to be unmethylated; whereas protein coding regions were largely (∼84%) methylated. Although CpG-rich and -poor promoters showed distinct methylation patterns, interestingly, a negative correlation between promoter methylation levels and gene transcription levels was consistently observed across promoters with high to low CpG densities. Importantly, we observed substantial interindividual variation in DNA methylation across the individual PBM methylomes and the pattern of this interindividual variation varied between different genomic features, with highly variable regions enriched for repetitive DNA elements. Furthermore, we observed a modest but significant excess (p < 2.2 × 10(-16)) of genes showing a negative correlation between interindividual promoter methylation and transcription levels. These significant genes were enriched in biological processes that are closely related to PBM functions, suggesting that alteration in DNA methylation is likely to be an important mechanism contributing to the interindividual variation in PBM function, and PBM-related phenotypic and disease-susceptibility variation in humans. CONCLUSION: This study represents a comprehensive analysis of the human PBM methylome and its interindividual variation. Our data provide a valuable resource for future epigenomic and multiomic studies, exploring biological and disease-related regulatory mechanisms in PBMs.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23750642',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 96 => array(
'id' => '1463',
'name' => 'Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation.',
'authors' => 'Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD',
'description' => 'Histone post-translational modifications regulate chromatin structure and function largely through interactions with effector proteins that often contain multiple histone-binding domains. While significant progress has been made characterizing individual effector domains, the role of paired domains and how they function in a combinatorial fashion within chromatin are poorly defined. Here we show that the linked tandem Tudor and plant homeodomain (PHD) of UHRF1 (ubiquitin-like PHD and RING finger domain-containing protein 1) operates as a functional unit in cells, providing a defined combinatorial readout of a heterochromatin signature within a single histone H3 tail that is essential for UHRF1-directed epigenetic inheritance of DNA methylation. These findings provide critical support for the "histone code" hypothesis, demonstrating that multivalent histone engagement plays a key role in driving a fundamental downstream biological event in chromatin.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23752590',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 97 => array(
'id' => '1449',
'name' => 'Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs.',
'authors' => 'Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R',
'description' => 'The presence of 5-methylcytidine (m(5)C) in tRNA and rRNA molecules of a wide variety of organisms was first observed more than 40 years ago. However, detection of this modification was limited to specific, abundant, RNA species, due to the usage of low-throughput methods. To obtain a high resolution, systematic, and comprehensive transcriptome-wide overview of m(5)C across the three domains of life, we used bisulfite treatment on total RNA from both gram positive (B. subtilis) and gram negative (E. coli) bacteria, an archaeon (S. solfataricus) and a eukaryote (S. cerevisiae), followed by massively parallel sequencing. We were able to recover most previously documented m(5)C sites on rRNA in the four organisms, and identified several novel sites in yeast and archaeal rRNAs. Our analyses also allowed quantification of methylated m(5)C positions in 64 tRNAs in yeast and archaea, revealing stoichiometric differences between the methylation patterns of these organisms. Molecules of tRNAs in which m(5)C was absent were also discovered. Intriguingly, we detected m(5)C sites within archaeal mRNAs, and identified a consensus motif of AUCGANGU that directs methylation in S. solfataricus. Our results, which were validated using m(5)C-specific RNA immunoprecipitation, provide the first evidence for mRNA modifications in archaea, suggesting that this mode of post-transcriptional regulation extends beyond the eukaryotic domain.',
'date' => '2013-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23825970',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 98 => array(
'id' => '1403',
'name' => 'Methyl donor supplementation blocks the adverse effects of maternal high fat diet on offspring physiology.',
'authors' => 'Carlin J, George R, Reyes TM',
'description' => 'Maternal consumption of a high fat diet during pregnancy increases the offspring risk for obesity. Using a mouse model, we have previously shown that maternal consumption of a high fat (60%) diet leads to global and gene specific decreases in DNA methylation in the brain of the offspring. The present experiments were designed to attempt to reverse this DNA hypomethylation through supplementation of the maternal diet with methyl donors, and to determine whether methyl donor supplementation could block or attenuate phenotypes associated with maternal consumption of a HF diet. Metabolic and behavioral (fat preference) outcomes were assessed in male and female adult offspring. Expression of the mu-opioid receptor and dopamine transporter mRNA, as well as global DNA methylation were measured in the brain. Supplementation of the maternal diet with methyl donors attenuated the development of some of the adverse effects seen in offspring from dams fed a high fat diet; including weight gain, increased fat preference (males), changes in CNS gene expression and global hypomethylation in the prefrontal cortex. Notable sex differences were observed. These findings identify the importance of balanced methylation status during pregnancy, particularly in the context of a maternal high fat diet, for optimal offspring outcome.',
'date' => '2013-05-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23658839',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 99 => array(
'id' => '1311',
'name' => 'Naive pluripotency is associated with global DNA hypomethylation.',
'authors' => 'Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, Surani MA, Hajkova P',
'description' => 'Naive pluripotent embryonic stem cells (ESCs) and embryonic germ cells (EGCs) are derived from the preimplantation epiblast and primordial germ cells (PGCs), respectively. We investigated whether differences exist between ESCs and EGCs, in view of their distinct developmental origins. PGCs are programmed to undergo global DNA demethylation; however, we find that EGCs and ESCs exhibit equivalent global DNA methylation levels. Inhibition of MEK and Gsk3b by 2i conditions leads to pronounced reduction in DNA methylation in both cell types. This is driven by Prdm14 and is associated with downregulation of Dnmt3a and Dnmt3b. However, genomic imprints are maintained in 2i, and we report derivation of EGCs with intact genomic imprints. Collectively, our findings establish that culture in 2i instills a naive pluripotent state with a distinctive epigenetic configuration that parallels molecular features observed in both the preimplantation epiblast and nascent PGCs.',
'date' => '2013-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23416945',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 100 => array(
'id' => '1290',
'name' => 'DNA methylation analysis in the intestinal epithelium-effect of cell separation on gene expression and methylation profile.',
'authors' => 'Jenke AC, Postberg J, Raine T, Nayak KM, Molitor M, Wirth S, Kaser A, Parkes M, Heuschkel RB, Orth V, Zilbauer M',
'description' => 'BACKGROUND: Epigenetic signatures are highly cell type specific. Separation of distinct cell populations is therefore desirable for all epigenetic studies. However, to date little information is available on whether separation protocols might influence epigenetic and/or gene expression signatures and hence might be less beneficial. We investigated the influence of two frequently used protocols to isolate intestinal epithelium cells (IECs) from 6 healthy individuals. MATERIALS AND METHODS: Epithelial cells were isolated from small bowel (i.e. terminal ileum) biopsies using EDTA/DTT and enzymatic release followed by magnetic bead sorting via EPCAM labeled microbeads. Effects on gene/mRNA expression were analyzed using a real time PCR based expression array. DNA methylation was assessed by pyrosequencing of bisulfite converted DNA and methylated DNA immunoprecipitation (MeDIP). RESULTS: While cell purity was >95% using both cell separation approaches, gene expression analysis revealed significantly higher mRNA levels of several inflammatory genes in EDTA/DTT when compared to enzymatically released cells. In contrast, DNA methylation of selected genes was less variable and only revealed subtle differences. Comparison of DNA methylation of the epithelial cell marker EPCAM in unseparated whole biopsy samples with separated epithelium (i.e. EPCAM positive and negative fraction) demonstrated significant differences in DNA methylation between all three tissue fractions indicating cell type specific methylation patterns can be masked in unseparated tissue samples. CONCLUSIONS: Taken together, our data highlight the importance of considering the potential effect of cell separation on gene expression as well as DNA methylation signatures. The decision to separate tissue samples will therefore depend on study design and specific separation protocols.',
'date' => '2013-02-08',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23409010',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 101 => array(
'id' => '1065',
'name' => 'Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation.',
'authors' => 'Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD',
'description' => 'A fundamental challenge in mammalian biology has been the elucidation of mechanisms linking DNA methylation and histone post-translational modifications. Human UHRF1 (ubiquitin-like PHD and RING finger domain-containing 1) has multiple domains that bind chromatin, and it is implicated genetically in the maintenance of DNA methylation. However, molecular mechanisms underlying DNA methylation regulation by UHRF1 are poorly defined. Here we show that UHRF1 association with methylated histone H3 Lys9 (H3K9) is required for DNA methylation maintenance. We further show that UHRF1 association with H3K9 methylation is insensitive to adjacent H3 S10 phosphorylation-a known mitotic 'phospho-methyl switch'. Notably, we demonstrate that UHRF1 mitotic chromatin association is necessary for DNA methylation maintenance through regulation of the stability of DNA methyltransferase-1. Collectively, our results define a previously unknown link between H3K9 methylation and the faithful epigenetic inheritance of DNA methylation, establishing a notable mitotic role for UHRF1 in this process.',
'date' => '2012-09-30',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23022729',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 102 => array(
'id' => '960',
'name' => 'Histone acetylation and DNA demethylation of T-cells result in an anaplastic large cell lymphoma-like phenotype.',
'authors' => 'Joosten M, Seitz V, Zimmermann K, Sommerfeld A, Berg E, Lenze D, Leser U, Stein H, Hummel M',
'description' => 'Background. A characteristic feature of anaplastic large cell lymphoma is the significant repression of the T-cell expression program despite its T-cell origin. The reasons for this down-regulation of T-cell phenotype are still unknown. Design and Methods. To elucidate whether epigenetic mechanisms are responsible for the loss of the T-cell phenotype, we treated anaplastic large cell lymphoma and T-cell lymphoma/leukemia cell lines (n=4, each) with epigenetic modifiers to evoke DNA demethylation and histone acetylation. Global gene expression data from treated and untreated cell lines were generated and selected differentially expressed genes were evaluated by real-time RT-PCR and Western Blot analysis. Additionally, histone H3 lysine 27 trimethylation was analyzed by chromatin immunoprecipitation. Results. Combined DNA demethylation and histone acetylation of anaplastic large cell lymphoma cells was not able to reconstitute their T-cell phenotype. Instead, the same treatment induced in T-cells (i) an up-regulation of anaplastic large cell lymphoma-characteristic genes (e.g. ID2, LGALS1, c-JUN) and (ii) an almost complete extinction of their T-cell phenotype including CD3, LCK and ZAP70. In addition, a suppressive trimethylation of histone H3 lysine 27 of important T-cell transcription factor genes (GATA3, LEF1, TCF1) was present in anaplastic large cell lymphoma cells, which is in line with their absence in primary tumour specimens as demonstrated by immunohistochemistry. Conclusions. Our data suggest that epigenetically activated suppressors (e.g. ID2) contribute to the down-regulation of the T-cell expression program in anaplastic large cell lymphoma, which is maintained by trimethylation of histone H3 lysine 27.',
'date' => '2012-08-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22899583',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 103 => array(
'id' => '389',
'name' => 'Growth Arrest and DNA-Damage-Inducible, Beta (GADD45b)-Mediated DNA Demethylation in Major Psychosis.',
'authors' => 'Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A',
'description' => 'Aberrant neocortical DNA methylation has been suggested to be a pathophysiological contributor to psychotic disorders. Recently, a growth arrest and DNA-damage-inducible, beta (GADD45b) protein-coordinated DNA demethylation pathway, utilizing cytidine deaminases and thymidine glycosylases, has been identified in the brain. We measured expression of several members of this pathway in parietal cortical samples from the Stanley Foundation Neuropathology Consortium (SFNC) cohort. We find an increase in GADD45b mRNA and protein in patients with psychosis. In immunohistochemistry experiments using samples from the Harvard Brain Tissue Resource Center, we report an increased number of GADD45b-stained cells in prefrontal cortical layers II, III, and V in psychotic patients. Brain-derived neurotrophic factor IX (BDNF IXabcd) was selected as a readout gene to determine the effects of GADD45b expression and promoter binding. We find that there is less GADD45b binding to the BDNF IXabcd promoter in psychotic subjects. Further, there is reduced BDNF IXabcd mRNA expression, and an increase in 5-methylcytosine and 5-hydroxymethylcytosine at its promoter. On the basis of these results, we conclude that GADD45b may be increased in psychosis compensatory to its inability to access gene promoter regions.',
'date' => '2012-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22048458',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 104 => array(
'id' => '409',
'name' => 'Epigenetic silencing mediated through activated PI3K/AKT signaling in breast cancer.',
'authors' => 'Zuo T, Liu TM, Lan X, Weng YI, Shen R, Gu F, Huang YW, Liyanarachchi S, Deatherage DE, Hsu PY, Taslim C, Ramaswamy B, Shapiro CL, Lin HJ, Cheng AS, Jin VX, Huang TH',
'description' => 'Trimethylation of histone 3 lysine 27 (H3K27me3) is a critical epigenetic mark for the maintenance of gene silencing. Additional accumulation of DNA methylation in target loci is thought to cooperatively support this epigenetic silencing during tumorigenesis. However, molecular mechanisms underlying the complex interplay between the two marks remain to be explored. Here we show that activation of PI3K/AKT signaling can be a trigger of this epigenetic processing at many downstream target genes. We also find that DNA methylation can be acquired at the same loci in cancer cells, thereby reinforcing permanent repression in those losing the H3K27me3 mark. Because of a link between PI3K/AKT signaling and epigenetic alterations, we conducted epigenetic therapies in conjunction with the signaling-targeted treatment. These combined treatments synergistically relieve gene silencing and suppress cancer cell growth in vitro and in xenografts. The new finding has important implications for improving targeted cancer therapies in the future.',
'date' => '2011-03-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/21216892',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 105 => array(
'id' => '412',
'name' => 'Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping.',
'authors' => 'Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, Weng YI, Deatherage DE, Chen Z, Pereira JS, Lopez R, Russo J, Wang Q, Lamartiniere CA, Nephew KP, Huang TH',
'description' => 'The current concept of epigenetic repression is based on one repressor unit corresponding to one silent gene. This notion, however, cannot adequately explain concurrent silencing of multiple loci observed in large chromosome regions. The long-range epigenetic silencing (LRES) can be a frequent occurrence throughout the human genome. To comprehensively characterize the influence of estrogen signaling on LRES, we analyzed transcriptome, methylome, and estrogen receptor alpha (ESR1)-binding datasets from normal breast epithelia and breast cancer cells. This "omics" approach uncovered 11 large repressive zones (range, 0.35 approximately 5.98 megabases), including a 14-gene cluster located on 16p11.2. In normal cells, estrogen signaling induced transient formation of multiple DNA loops in the 16p11.2 region by bringing 14 distant loci to focal ESR1-docking sites for coordinate repression. However, the plasticity of this free DNA movement was reduced in breast cancer cells. Together with the acquisition of DNA methylation and repressive chromatin modifications at the 16p11.2 loci, an inflexible DNA scaffold may be a novel determinant used by breast cancer cells to reinforce estrogen-mediated repression.',
'date' => '2010-06-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/20442245',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 106 => array(
'id' => '976',
'name' => 'Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells.',
'authors' => 'Borgel J, Guibert S, Weber M.',
'description' => 'Methylated DNA immunoprecipitation (MeDIP) is an immunocapturing approach for unbiased enrichment of DNA that is methylated on cytosines. The principle is that genomic DNA is randomly sheared by sonication and immunoprecipitated with an antibody that specifically recognizes 5-methylcytidine (5mC), which can be combined with PCR or high-throughput analysis (microarrays, deep sequencing). The MeDIP technique has been originally used to generate DNA methylation profiles on a genome scale in mammals and plants. Here we provide an optimized version of the MeDIP protocol suitable for low amounts of DNA, which can be used to study DNA methylation in cellular populations available in small quantities.',
'date' => '0000-00-00',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22907495',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '120',
'name' => '5-mC Antibody - clone 33D3 SDS US en',
'language' => 'en',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-US-en-GHS_2_0.pdf',
'countries' => 'US',
'modified' => '2020-03-13 15:47:06',
'created' => '2020-03-13 15:47:06',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '118',
'name' => '5-mC Antibody - clone 33D3 SDS GB en',
'language' => 'en',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-GB-en-GHS_2_0.pdf',
'countries' => 'GB',
'modified' => '2020-03-13 15:44:22',
'created' => '2020-03-13 15:44:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '116',
'name' => '5-mC Antibody - clone 33D3 SDS ES es',
'language' => 'es',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-ES-es-GHS_2_0.pdf',
'countries' => 'ES',
'modified' => '2020-03-13 15:43:01',
'created' => '2020-03-13 15:43:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '115',
'name' => '5-mC Antibody - clone 33D3 SDS DE de',
'language' => 'de',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-DE-de-GHS_2_0.pdf',
'countries' => 'DE',
'modified' => '2020-03-13 15:42:22',
'created' => '2020-03-13 15:42:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '119',
'name' => '5-mC Antibody - clone 33D3 SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-JP-ja-GHS_4_0.pdf',
'countries' => 'JP',
'modified' => '2020-03-13 15:44:56',
'created' => '2020-03-13 15:44:56',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '114',
'name' => '5-mC Antibody - clone 33D3 SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-03-13 15:41:00',
'created' => '2020-03-13 15:41:00',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '113',
'name' => '5-mC Antibody - clone 33D3 SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-BE-fr-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-03-13 15:40:12',
'created' => '2020-03-13 15:40:12',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '117',
'name' => '5-mC Antibody - clone 33D3 SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-03-13 15:43:48',
'created' => '2020-03-13 15:43:48',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = true
$other_formats = array(
(int) 0 => array(
'id' => '1980',
'antibody_id' => '630',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>Monoclonal antibody raised in mouse against </span><b>5-mC</b><span><span> </span>(</span><b>5-methylcytosine</b><span>) conjugated to ovalbumine (</span><b>33D3 clone</b><span>).</span></p>',
'label1' => 'Validation Data',
'info1' => '<div class="row">
<div class="small-12 columns">
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-A.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="173" /></p>
<p class="text-center"><img src="https://www.diagenode.com/img/product/antibodies/C15200081_ChIPSeq-B.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP-seq" caption="false" width="886" height="184" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 1. MeDIP-seq with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> Genomic DNA from E14 ES cells was sheared with the Bioruptor® to generate random fragments (size range 300 to 700 bp). One µg of the fragmented DNA was ligated to Illumina adapters and the resulting DNA was used for a standard MeDIP assay, using 2 µg of the Diagenode monoclonal against 5-mC (Cat. No. C15200081). After recovery of the methylated DNA, Illumina sequencing libraries were generated and sequenced on an Illumina Genome Analyzer according to the manufacturer’s instructions. Figure 1A and 1B show Genome browser views of CA simple repeat elements with read distributions specific for 5-mC at 2 gene locations (SigleC15 and Mfsd4). Visual inspection of the peak profiles in a genome browser reveals high enrichment of CA simple repeats in affinity-enriched methylated fragments after MeDIP with the Diagenode 5-mC monoclonal antibody.</small></p>
</div>
</div>
<div class="row">
<div class="small-5 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_medip.png" alt="5-mC (5-methylcytosine) Antibody validated in MeDIP" caption="false" width="355" height="372" /></p>
</div>
<div class="small-7 columns">
<p><small><strong>Figure 2. MeDIP results obtained with the Diagenode monoclonal antibody directed against 5-mC</strong><br /> MeDIP (Methylated DNA immunoprecipitation) was performed on 1 µg fragmented human genomic DNA using 0.2 µg of the Diagenode monoclonal antibody against 5-mC (cat. No. C15200081) and the MagMeDIP Kit (cat. No. C02010021). The fragmented DNA was spiked with the internal controls present in the kit (methylated DNA (meDNA) as a positive and unmethylated DNA (unDNA) as a negative control) prior to performing the IP. QPCR was performed with optimized primer sets, included in the kit, specific for the methylated and unmethylated DNA controls, and for a known methylated (TSH2B) and unmethylated (GAPDH) genomic region. Figure 2 shows the recovery expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-3 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200081_Dotblot.png" alt=" 5-mC (5-methylcytosine) Antibody validated in dot blot" caption="false" width="201" height="196" /></p>
</div>
<div class="small-9 columns">
<p><small><strong>Figure 3. Dot blot analysis using the Diagenode monoclonal antibody directed against 5-mC</strong><br />To demonstrate the specificity of the Diagenode antibody against 5-mC (cat. No. C15200081), a Dot blot analysis was performed using the hmC, mC and C controls from the Diagenode “5-hmC, 5-mC & cytosine DNA Standard Pack” (cat. No. C02040010). One hundred to 4 ng (equivalent of 5 to 0.2 pmol of C-bases) of the controls were spotted on a membrane. Figure 3 shows a high specificity of the antibody for the methylated control.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_IF1.png" alt="5-mC (5-methylcytosine) Antibody for immunofluorescence" height="121" width="500" caption="false" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 4. Immunofluorescence using the Diagenode monoclonal antibody directed against 5-mC</strong><br />HeLa cells were stained with the Diagenode antibody against 5-mC (Cat. No. C15200081) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 1% BSA. The cells were immunofluorescently labelled with the 5-mC antibody (middle) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The left panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>
<!--
<div class="row">
<div class="small-12 columns"><center><img src="https://www.diagenode.com/img/product/antibodies/C15200081_SPR.png" alt="5-methylcytosine (5-mC) Antibody" surface="" plasmon="" resonance="" caption="false" width="700" height="372" /></center></div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong>Figure 5. Surface plasmon resonance (SPR) analysis of the the Diagenode monoclonal antibody directed against 5-mC</strong><br />A synthesized biotin-labeled 5-mC conjugate was immobilized on a CM4 BIAcore sensorchip (GE Healthcare, France). Briefly, two flowcells were prepared by sequential injections of EDC/NHS, streptavidin, and ethanolamine. One of these flowcells served as negative control (biotinylated spacer without 5-mC), while biotinylated 5-mC conjugate was injected in the other one, to get an immobilization level of 55 response units (RU). All SPR experiments were performed, using HBS-N buffer (10 mM HEPES,150 mM NaCl, pH 7.4), at a flow rate of 5 µl/min. Interaction assays involved injections of 2 different dilutions of the Diagenode 5-mC monoclonal antibody (Cat. No. C15200081) over the biotinylated 5-mC conjugate and negative control surfaces, followed by a 3 min washing step with HBS-N buffer to allow dissociation of the complexes formed. At the end of each cycle, the streptavidin surface was regenerated by injection of 0.1M citric acid (pH=3).</small></p>
<p><small>The sensorgrams correspond to the biotinylated 5-mC conjugate surface signal subtracted with the negative control. Data from the sensorgrams that reached binding equilibrium were used for Scatchard analysis. The value of the dissociation constant (kd) obtained by global fitting and 1:1 Langmuir model is 65 nM.</small></p>
</div>
</div>-->',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15200081-100',
'old_catalog_number' => 'MAb-081-100',
'sf_code' => 'C15200081-D001-000526',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '505',
'price_USD' => '575',
'price_GBP' => '450',
'price_JPY' => '79110',
'price_CNY' => '0',
'price_AUD' => '1438',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'October 27, 2020',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-100-ug-50-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081) | Diagenode',
'meta_keywords' => '5-methylcytosine (5-mC),monoclonal antibody,Methylated DNA Immunoprecipitation',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2023-05-17 10:08:33',
'created' => '2015-06-29 14:08:20'
),
(int) 1 => array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
)
$pro = array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010034</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1885" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1885" id="CartAdd/1885Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1885" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto hMeDIP kit x16 (monoclonal mouse antibody)</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns" data-reveal-id="cartModal-1885" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto hMeDIP kit x16 (monoclonal mouse antibody)</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/magmedip-kit-x48-48-rxns"><img src="/img/product/kits/C02010021-magmedip-qpcr.jpg" alt="MagMeDIP qPCR Kit box" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010021</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1880" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1880" id="CartAdd/1880Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1880" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MagMeDIP Kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MagMeDIP Kit',
'C02010021',
'750',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MagMeDIP Kit',
'C02010021',
'750',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="magmedip-kit-x48-48-rxns" data-reveal-id="cartModal-1880" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MagMeDIP Kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/auto-methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020011</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1888" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1888" id="CartAdd/1888Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1888" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto MethylCap kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1888" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto MethylCap kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1887" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1887" id="CartAdd/1887Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1887" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MethylCap kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MethylCap kit',
'C02020010',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MethylCap kit',
'C02020010',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1887" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MethylCap kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/premium-bisulfite-kit-50-rxns"><img src="/img/grey-logo.jpg" alt="default alt" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02030030</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1892" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/en/carts/add/1892" id="CartAdd/1892Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1892" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>Add <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Premium Bisulfite kit</strong> to my shopping cart.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Premium Bisulfite kit',
'C02030030',
'240',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">Checkout</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Premium Bisulfite kit',
'C02030030',
'240',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">Keep shopping</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="premium-bisulfite-kit-50-rxns" data-reveal-id="cartModal-1892" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Premium Bisulfite kit</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1892',
'antibody_id' => null,
'name' => 'Premium Bisulfite kit',
'description' => '<p style="text-align: center;"><a href="https://www.diagenode.com/files/products/kits/Premium_Bisulfite_kit_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p style="text-align: center;"><strong>Make your Bisulfite conversion now in only 60 minutes !</strong></p>
<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C02030030',
'old_catalog_number' => '',
'sf_code' => 'C02030030-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'premium-bisulfite-kit-50-rxns',
'meta_title' => 'Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Premium Bisulfite kit',
'modified' => '2023-04-20 16:13:50',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '271',
'product_id' => '1979',
'related_id' => '1892'
),
'Image' => array()
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ' <span style="color:#CCC">(MAb-081-500)</span>'
$country_code = 'US'
$other_format = array(
'id' => '1981',
'antibody_id' => '62',
'name' => '5-methylcytosine (5-mC) Antibody - clone 33D3',
'description' => '<p><span>The <strong>5-methylcytosine</strong> antibody (<strong>clone 33D3</strong>) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span><br /><br /><strong><span>Diagenode is the exclusive worldwide source of genuine 33D3 clone!</span></strong></p>',
'label1' => 'Validation Data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '500 µg',
'catalog_number' => 'C15200081-500',
'old_catalog_number' => 'MAb-081-500',
'sf_code' => 'C15200081-D001-000527',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '1770',
'price_USD' => '1950',
'price_GBP' => '1585',
'price_JPY' => '277270',
'price_CNY' => '',
'price_AUD' => '4875',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '0000-00-00',
'slug' => '5-mc-monoclonal-antibody-33d3-premium-500-ug-250-ul',
'meta_title' => '5-methylcytosine (5-mC) Antibody - clone 33D3 (C15200081-500) | Diagenode',
'meta_keywords' => '',
'meta_description' => '5-methylcytosine (5-mC) Monoclonal Antibody, clone 33D3 validated in MeDIP-seq, MeDIP, DB and IF. Batch-specific data available on the website. Sample size available.',
'modified' => '2022-06-29 13:59:03',
'created' => '2015-06-29 14:08:20',
'ProductsGroup' => array(
'id' => '6',
'product_id' => '1981',
'group_id' => '4'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
'id' => '1914',
'product_id' => '1979',
'application_id' => '29'
)
)
$slugs = array(
(int) 0 => 'immunofluorescence'
)
$applications = array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'locale' => 'eng'
)
$description = '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>'
$name = 'IF'
$document = array(
'id' => '1118',
'name' => 'Datasheet 5-mC33D3 C15200081-10',
'description' => '<p><span>The 5-methylcytosine antibody (clone 33D3) is the most published and widely used antibody for DNA methylation analysis. It has been validated for Methylated DNA Immunoprecipitation (MeDIP-seq, MeDIP-on-chip), Immunofluorescence and Dot blot. </span></p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_5-mC33D3_C15200081-10.pdf',
'slug' => 'datasheet-5-mc33d3-c15200081-10',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-12-17 10:55:09',
'created' => '2020-12-17 10:55:09',
'ProductsDocument' => array(
'id' => '3063',
'product_id' => '1979',
'document_id' => '1118'
)
)
$sds = array(
'id' => '117',
'name' => '5-mC Antibody - clone 33D3 SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/5-mC/SDS-C15200081-5-methylcytosine_5-mC_Antibody_-_clone_33D3-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-03-13 15:43:48',
'created' => '2020-03-13 15:43:48',
'ProductsSafetySheet' => array(
'id' => '231',
'product_id' => '1979',
'safety_sheet_id' => '117'
)
)
$publication = array(
'id' => '976',
'name' => 'Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells.',
'authors' => 'Borgel J, Guibert S, Weber M.',
'description' => 'Methylated DNA immunoprecipitation (MeDIP) is an immunocapturing approach for unbiased enrichment of DNA that is methylated on cytosines. The principle is that genomic DNA is randomly sheared by sonication and immunoprecipitated with an antibody that specifically recognizes 5-methylcytidine (5mC), which can be combined with PCR or high-throughput analysis (microarrays, deep sequencing). The MeDIP technique has been originally used to generate DNA methylation profiles on a genome scale in mammals and plants. Here we provide an optimized version of the MeDIP protocol suitable for low amounts of DNA, which can be used to study DNA methylation in cellular populations available in small quantities.',
'date' => '0000-00-00',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22907495',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
'id' => '711',
'product_id' => '1979',
'publication_id' => '976'
)
)
$externalLink = ' <a href="http://www.ncbi.nlm.nih.gov/pubmed/22907495" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
×