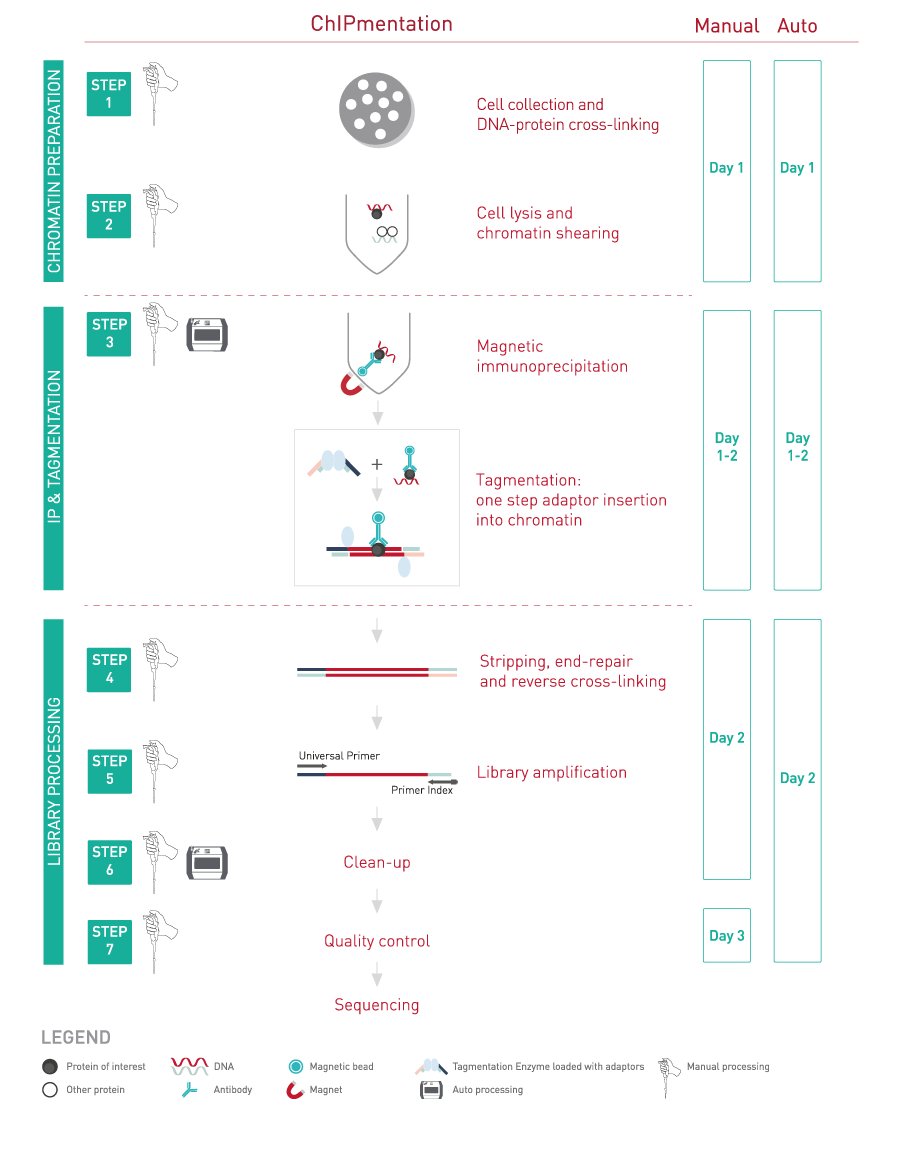
Diagenode’s ChIPmentation technology, based on tagmentation, enables the integration of library preparation during ChIP itself using transposase and sequencing-compatible adaptors. Unlike standard library preparation techniques that require multi-step ligation, ChIPmentation incorporates an easier and shorter protocol.
Different protocols have been optimized bringing the solution for different needs:
| Cat. No. | Product | Format | Price | |
|---|---|---|---|---|
| C01011011 |
µChIPmentation Kit for Histones Download the Manual The Diagenode µChIPmentation Kit for Histones is optimized to perform ChIP-seq on as little as 10.000 cells from cell fixation to ... |
24 rxns | $2,455.00 | |
| C01011032 |
24 SI for Tagmented libraries The 24 SI for tagmented libraries includes 24 single primer indexes for multiplexing up to 24 samples. The primer indexes of the 24 SI Tagmented libraries ... |
24 SI | $195.00 | |
| C01011000 |
Auto ChIPmentation Kit for Histones - Replaced by C01011009 Contact us This product must be used with the IP-Star Compact Automated System. Diagenode’s latest technology for histone ChIP-seq, ... |
24 rxns | ||
| C01011010 |
ChIPmentation Kit for Histones - Replaced by C01011009 Contact us Difficult and challenging histone ChIP-seq is now solved. Diagenode’s latest technology for histone ChIP-seq, ChIPmentation ... |
24 rxns | ||
| C01011030 |
TAG Kit for ChIPmentation The TAG Kit for ChIPmentation offers an optimized ChIP-seq library preparation solution based on tagmentation. This kit includes reagents for ... |
24 rxns | $1,075.00 |