The 24 UDI (Unique dual indexes) for tagmented libraries – Set I is compatible with any tagmentation-based library preparation protocols, such as ChIPmentation, ATAC-seq or CUT&Tag technologies.
The 24 UDI for tagmented libraries provides combinations of barcodes where each barcode is uniquely attributed to one sample. This is a great tool to identify mistakes during index sequencing. A phenomenon, known as index hopping, can lead to misattribution of some reads to the wrong sample. This is particularly frequent with the NovaSeq6000, and thus the use of Unique Dual Indexing (UDI) is highly recommended when using this sequencer.
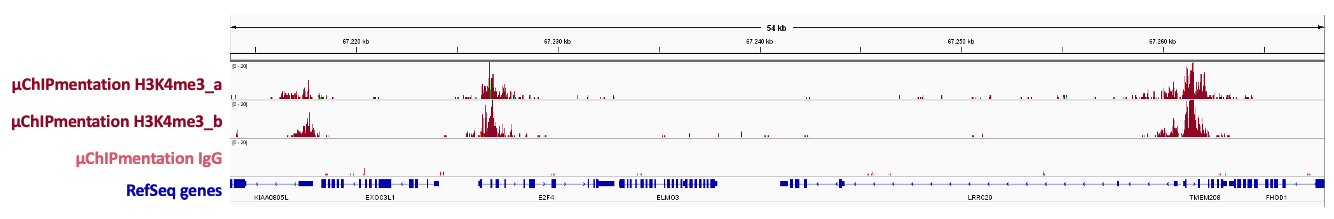
Figure 1. Sequencing profiles of µChIPmentation libraries generated with 24 UDI for Tagmented libraries Chromatin preparation and immunoprecipitation have been performed on 10.000 cells using the µChIPmentation Kit for Histones (Cat. No. C01011011) and 24 UDI for Tagmented libraries – Set I (Cat. No. Cat. No. C01011034) using K562 cells. The Diagenode antibodies targeting H3K4me3 (Cat. No. C15410003) and rabbit IgG (Cat. No. C15410206) have been used.



