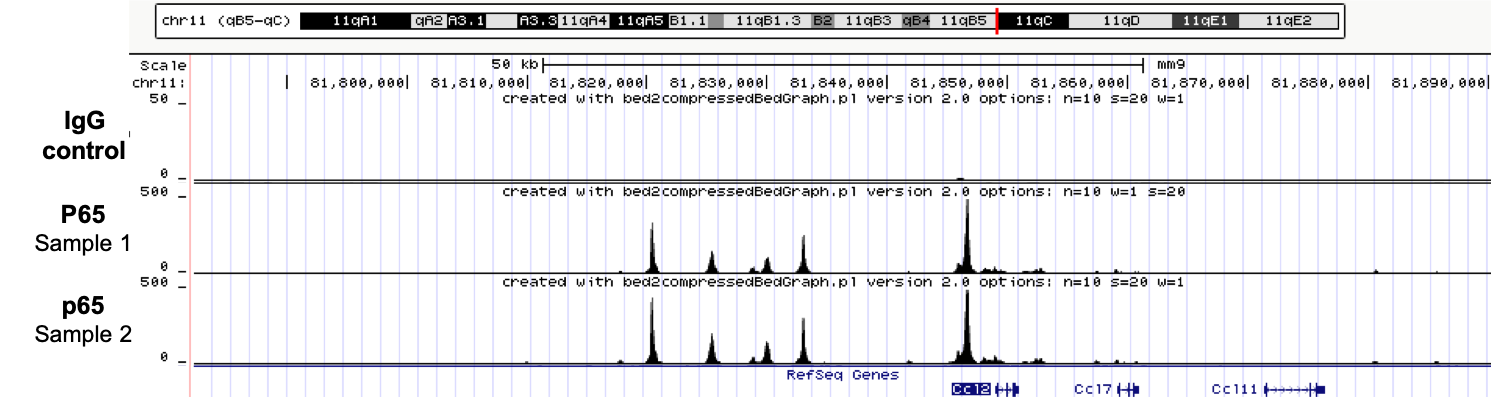
Comparing to classical ChIP-seq, ChIPmentation enables:
- An easier protocol
- Faster time to results
- Flexibility – the TAG Kit for ChIPmentation allows you to perform ChIPmentation with any ChIP protocol (based on magnetic beads)
ChIPmentation on histone marks using TAG KIT for ChIPmentation
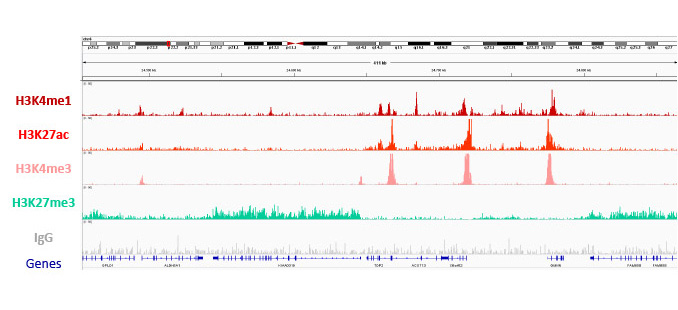
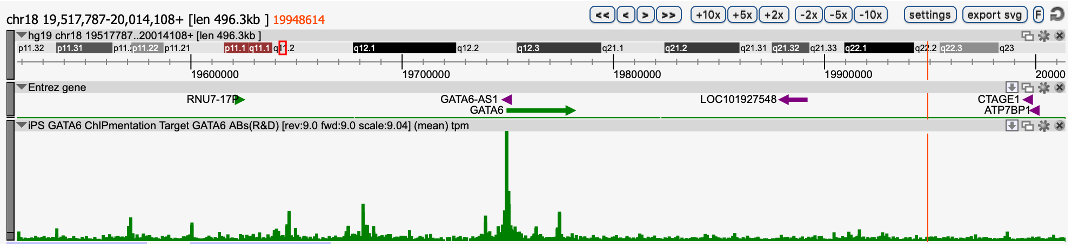
ChIPmentation sequencing results for four histone marks. Chromatin preparation has been performed on 7M K562 cells using the Auto iDeal ChIP-seq Kit for Histones (Cat. No. C01010057), TAG Kit for ChIPmentation (Cat. No. C01011030) and 24 SI for ChIPmentation (Cat. No. C01011031)*. Diluted chromatin from 1,000,000 cells was used for the immunoprecipitation with the Diagenode antibodies targeting H3K4me1 (Cat. No. C15410194), H3K27me3 (Cat. No. C15410195), H3K27ac (Cat. No. C15410196) and IgG (Cat. No. C15410206). The experiment was performed on the IP-Star® Compact Automated System.
*The Auto iDeal ChIP-seq for Histones (C01010057), TAG Kit for ChIPmentation (C01011030) and 24 SI for ChIPmentation (C01011031) form together the Auto ChIPmentation Kit for Histones* (C01011000).
ChIPmentation on non-histone proteins using TAG Kit for ChIPmentation
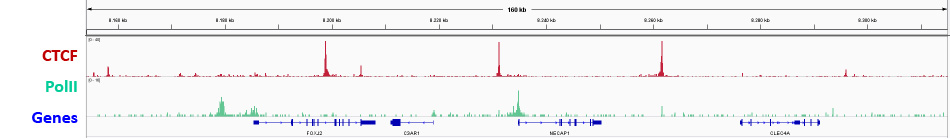
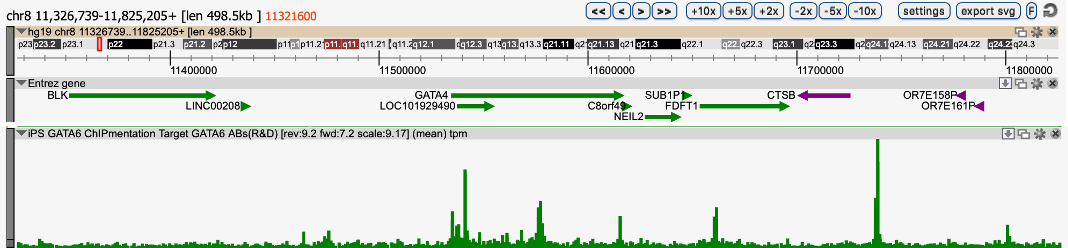
ChIPmentation sequencing results for two non histone factors. Chromatin preparation has been performed on 25M K562 cells using the iDeal ChIP-seq kit for TFs (Cat. No. C01010055). Diluted chromatin from 1,000,000 or 4,000,000 cells was used for the immunoprecipitation with the Diagenode antibodies targeting CTCF (Cat. No. C15410210) and RNA PolII (Cat. No. C15100055 ) respectively. The library preparation was performed with the TAG Kit for ChIPmentation (Cat. No. C01011030) and 24 SI for ChIPmentation (Cat. No. C01011031).