For over 15 years Diagenode has been the most trusted supplier for epigenetics-focused antibodies. We understand that high quality is crucial for the success of experiments, so we emphasize strict quality standards and the actual validation for all of our antibodies in applications such ChIP and ChIP-seq. Our antibodies are produced and validated following the ChIP-seq guidelines defined by major epigenetic consortia including Blueprint and ENCODE. Recently our standard antibody QC has been enriched by CUT&Tag validation enabling to select the antibodies with high performance in this assay.
A.
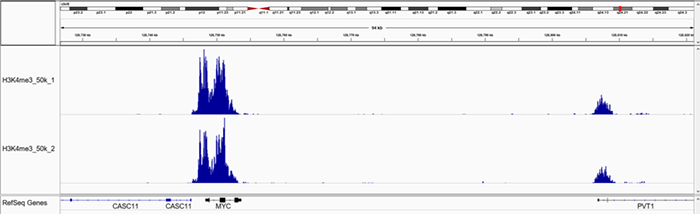
B.
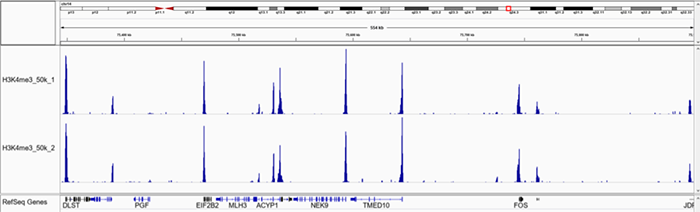
C.
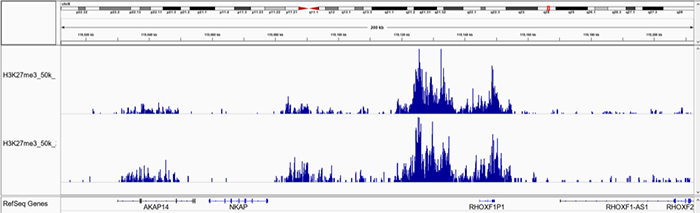
D.
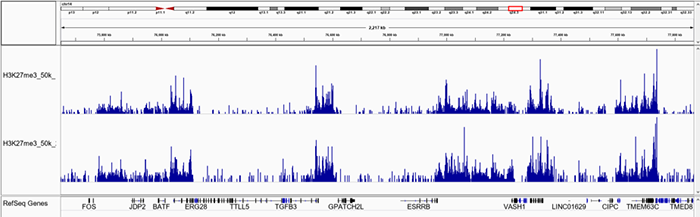
E.
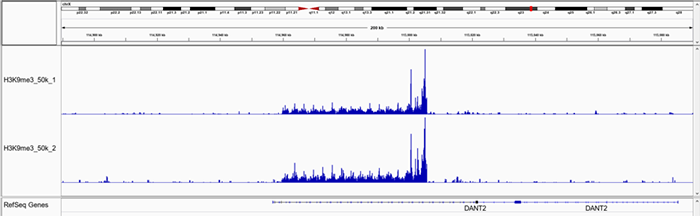
F.
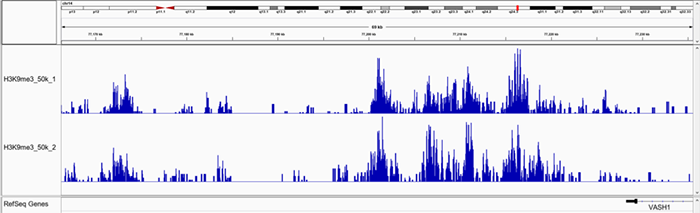
Figure 1. Representative screenshot for H3K4me3 (A and B), H3K27me3 (C and D), H3K9me3 (E and F) data obtained using iDeal CUT&Tag kit for Histones (Cat. No. C01070020) and Antibody package for Histones anti-rabbit (Cat. No. C01070022) at selected loci. CUT&Tag was performed using 50,000 K562 cells and Diagenode H3K4me3 polyclonal CUT&Tag antibody (Cat. No. C15410003), H3K27me3 polyclonal CUT&Tag grade antibody (Cat. No. C15410195) and H3K9me3 poyclonal CUT&Tag grade antibody (Cat. No. C15410193).
A.
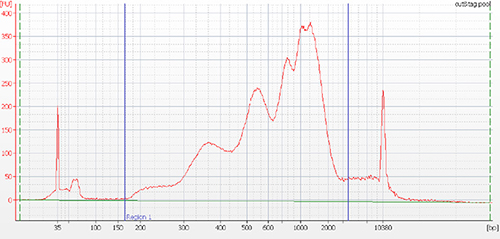
B.
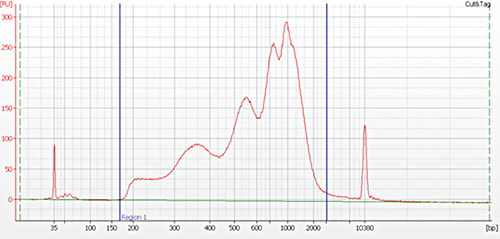
Figure 2. Typical library profile generated by CUT&Tag protocol. Agilent BioAnalyzer trace of pooled libraries derived from H3K4me3, H3K27me3 and H3K9me3 CUT&Tag experiments performed using 50,000 K562 cells (A) and from H3K27me3 and H3K9me3 CUT&Tag experiments using 25 000, 10 000 and 1 000 K562 cells (B). The following Diagenode antibodies were used: H3K4me3 polyclonal CUT&Tag antibody (Cat. No. C15410003), H3K27me3 polyclonal CUT&Tag grade antibody (Cat. No. C15410195) and H3K9me3 polyclonal CUT&Tag grade antibody (Cat. No. C15410193).