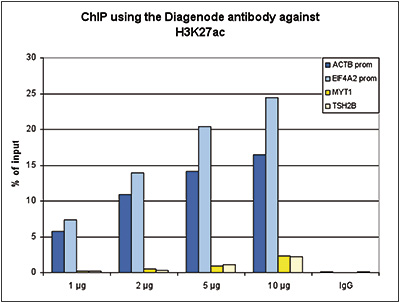
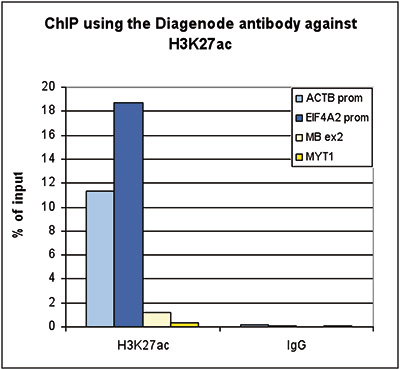
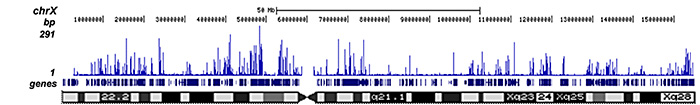
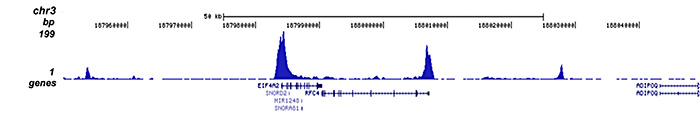
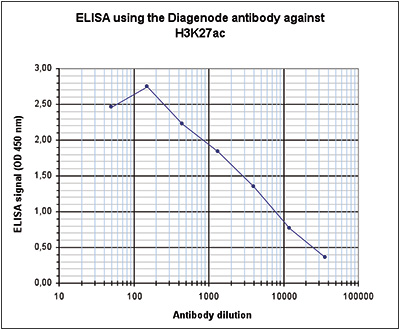
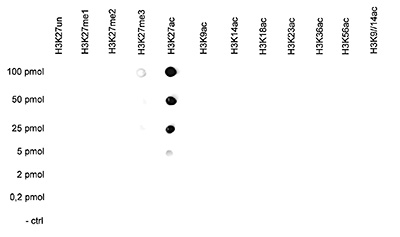
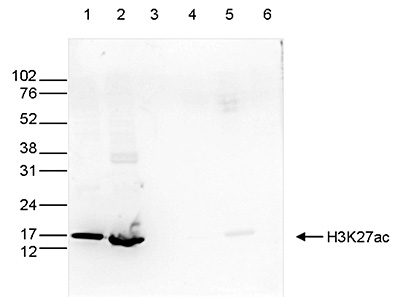
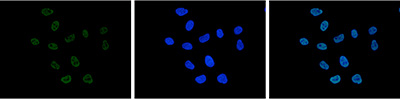
How to properly cite our product/service in your work We strongly recommend using this: H3K27ac Antibody (Hologic Diagenode Cat# C15410174 Lot# A7071-001P). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Liebenberg syndrome severity arises from variations in Pitx1 locus topology and proportion of ectopically transcribing cells
Bompadre, Olimpia et al.
Enhancer hijacking, a common cause of gene misregulation linked to disease, occurs when non-matching enhancers and promoters interact ectopically due to genetic alterations. While the concept of enhancer hijacking is well understood, the reasons behind the variation in phenotypic severity remain unexplored. In t... |
Incomplete transcriptional dosage compensation of vertebrate sexchromosomes is balanced by post-transcriptional compensation
Lister N. C. et al.
Heteromorphic sex chromosomes (XY or ZW) present problems of gene dosage imbalance between the sexes, and with the autosomes. Mammalian X chromosome inactivation was long thought to imply a critical need for dosage compensation in vertebrates. However, the universal importance of sex chromosome dosage compensation w... |
Gene Regulatory Interactions at Lamina-Associated Domains
Madsen-Østerbye J. et al.
The nuclear lamina provides a repressive chromatin environment at the nuclear periphery. However, whereas most genes in lamina-associated domains (LADs) are inactive, over ten percent reside in local euchromatic contexts and are expressed. How these genes are regulated and whether they are able to interact with regu... |
Repression and 3D-restructuring resolves regulatory conflicts inevolutionarily rearranged genomes.
Ringel A. et al.
Regulatory landscapes drive complex developmental gene expression, but it remains unclear how their integrity is maintained when incorporating novel genes and functions during evolution. Here, we investigated how a placental mammal-specific gene, Zfp42, emerged in an ancient vertebrate topologically associated domai... |
Epigenetic remodeling of downstream enhancer regions is linked toselective expression of the IL1F10 gene in differentiated humankeratinocytes.
Talabot-Ayer D. et al.
Interleukin (IL)-38, encoded by the IL1F10 gene, is a member of the IL-1 family of cytokines. IL-38 is constitutively expressed in epithelia in healthy humans, and in particular in epidermal keratinocytes in the skin. IL-38 expression is closely correlated with keratinocyte differentiation. The aim of this study was... |
HOTAIR interacts with PRC2 complex regulating the regional preadipocytetranscriptome and human fat distribution.
Kuo Feng-Chih et al.
Mechanisms governing regional human adipose tissue (AT) development remain undefined. Here, we show that the long non-coding RNA HOTAIR (HOX transcript antisense RNA) is exclusively expressed in gluteofemoral AT, where it is essential for adipocyte development. We find that HOTAIR interacts with polycomb repressive ... |
Nox4 promotes endothelial differentiation through chromatin remodeling.
Hahner F. et al.
RATIONALE: Nox4 is a constitutively active NADPH oxidase that constantly produces low levels of HO. Thereby, Nox4 contributes to cell homeostasis and long-term processes, such as differentiation. The high expression of Nox4 seen in endothelial cells contrasts with the low abundance of Nox4 in stem cells, which are a... |
Local euchromatin enrichment in lamina-associated domains anticipatestheir repositioning in the adipogenic lineage.
Madsen-Østerbye J. et al.
BACKGROUND: Interactions of chromatin with the nuclear lamina via lamina-associated domains (LADs) confer structural stability to the genome. The dynamics of positioning of LADs during differentiation, and how LADs impinge on developmental gene expression, remains, however, elusive. RESULTS: We examined changes in t... |
Screening of ETO2-GLIS2-induced Super Enhancers identifiestargetable cooperative dependencies in acute megakaryoblastic leukemia.
Benbarche S. et al.
Super Enhancers (SEs) are clusters of regulatory elements associated with cell identity and disease. However, whether these elements are induced by oncogenes and can regulate gene modules cooperating for cancer cell transformation or maintenance remains elusive. To address this question, we conducted a genome-wide C... |
Loss of KMT2C reprograms the epigenomic landscape in hPSCsresulting in NODAL overexpression and a failure of hemogenic endotheliumspecification.
Maurya Shailendra et al.
Germline or somatic variation in the family of KMT2 lysine methyltransferases have been associated with a variety of congenital disorders and cancers. Notably, -fusions are prevalent in 70\% of infant leukaemias but fail to phenocopy short latency leukaemogenesis in mammalian models, suggesting additional factors ar... |
SETD2-mediated epigenetic regulation of noncanonical Wnt5A duringosteoclastogenesis
Deb M. et al.
Graphic abstract To define the role of SETD2 in the WNT5a signaling in the context of osteoclastogenesis, we exploited two different models: in vitro osteoclast differentiation, and K/BxN serum-induced arthritis model. We found that SETD2 and WNT5a were upregulated during osteoclast differentiation and after inducti... |
Activated Histone Acetyltransferase p300/CBP-Related SignallingPathways Mediate Up-Regulation of NADPH Oxidase,Inflammation, and Fibrosis in Diabetic Kidney
Alexandra-Gela Lazar et al.
Accumulating evidence implicates the histone acetylation-based epigenetic mechanisms in the pathoetiology of diabetes-associated micro-/macrovascular complications. Diabetic kidney disease (DKD) is a progressive chronic inflammatory microvascular disorder ultimately leading to glomerulosclerosis and kidney failure. ... |
Cell-specific alterations inPitx1regulatory landscape activation caused bythe loss of a single enhancer
Rouco, R. et al.
Most developmental genes rely on multiple transcriptional enhancers for their accurate expression during embryogenesis. Because enhancers may have partially redundant activities, the loss of one of them often leads to a partial loss of gene expression and concurrent moderate phenotypic outcome, if any. While such a ... |
Functional annotations of three domestic animal genomes provide vitalresources for comparative and agricultural research.
Kern C. et al.
Gene regulatory elements are central drivers of phenotypic variation and thus of critical importance towards understanding the genetics of complex traits. The Functional Annotation of Animal Genomes consortium was formed to collaboratively annotate the functional elements in animal genomes, starting with domesticate... |
A Tumor Suppressor Enhancer of PTEN in T-cell development and leukemia
L. Tottone at al.
Long-range oncogenic enhancers play an important role in cancer. Yet, whether similar regulation of tumor suppressor genes is relevant remains unclear. Loss of expression of PTEN is associated with the pathogenesis of various cancers, including T-cell leukemia (T-ALL). Here, we identify a highly conserved distal enh... |
EZH2 and KDM6B Expressions Are Associated with Specific EpigeneticSignatures during EMT in Non Small Cell Lung Carcinomas.
Lachat C. et al.
The role of Epigenetics in Epithelial Mesenchymal Transition (EMT) has recently emerged. Two epigenetic enzymes with paradoxical roles have previously been associated to EMT, EZH2 (Enhancer of Zeste 2 Polycomb Repressive Complex 2 (PRC2) Subunit), a lysine methyltranserase able to add the H3K27me3 mark, and the hist... |
Enhancer hijacking determines extrachromosomal circular MYCN ampliconarchitecture in neuroblastoma.
Helmsauer, Konstantin and Valieva, Maria E and Ali, Salaheddine andChamorro González, Rocío and Schöpflin, Robert and Röefzaad, Claudiaand Bei, Yi and Dorado Garcia, Heathcliff and Rodriguez-Fos, Elias andPuiggròs, Montserrat and Kasack, Katharina and
MYCN amplification drives one in six cases of neuroblastoma. The supernumerary gene copies are commonly found on highly rearranged, extrachromosomal circular DNA (ecDNA). The exact amplicon structure has not been described thus far and the functional relevance of its rearrangements is unknown. Here, we analyze the M... |
ΔNp63 is a pioneer factor that binds inaccessible chromatin and elicitchromatin remodeling
Yu X. et al.
Background: ΔNp63 is a master transcriptional regulator playing critical roles in epidermal development and other cellular processes. Recent studies suggest that ΔNp63 functions as a pioneer factor that can target its binding sites within inaccessible chromatin and induce chromatin remodeling. Methods: I... |
Distinct and temporary-restricted epigenetic mechanisms regulate human αβ and γδ T cell development
Roels J, Kuchmiy A, De Decker M, et al.
The development of TCRαβ and TCRγδ T cells comprises a step-wise process in which regulatory events control differentiation and lineage outcome. To clarify these mechanisms, we employed RNA-sequencing, ATAC-sequencing and ChIPmentation on well-defined thymocyte subsets that represent the conti... |
HDAC3 functions as a positive regulator in Notch signal transduction.
Ferrante F, Giaimo BD, Bartkuhn M, Zimmermann T, Close V, Mertens D, Nist A, Stiewe T, Meier-Soelch J, Kracht M, Just S, Klöble P, Oswald F, Borggrefe T
Aberrant Notch signaling plays a pivotal role in T-cell acute lymphoblastic leukemia (T-ALL) and chronic lymphocytic leukemia (CLL). Amplitude and duration of the Notch response is controlled by ubiquitin-dependent proteasomal degradation of the Notch1 intracellular domain (NICD1), a hallmark of the leukemogenic pro... |
Long non-coding RNA uc.291 controls epithelial differentiation by interfering with the ACTL6A/BAF complex.
Panatta E, Lena AM, Mancini M, Smirnov A, Marini A, Delli Ponti R, Botta-Orfila T, Tartaglia GG, Mauriello A, Zhang X, Calin GA, Melino G, Candi E
The mechanisms that regulate the switch between epidermal progenitor state and differentiation are not fully understood. Recent findings indicate that the chromatin remodelling BAF complex (Brg1-associated factor complex or SWI/SNF complex) and the transcription factor p63 mutually recruit one another to open chroma... |
Distinct IL-1α-responsive enhancers promote acute and coordinated changes in chromatin topology in a hierarchical manner.
Weiterer SS, Meier-Soelch J, Georgomanolis T, Mizi A, Beyerlein A, Weiser H, Brant L, Mayr-Buro C, Jurida L, Beuerlein K, Müller H, Weber A, Tenekeci U, Dittrich-Breiholz O, Bartkuhn M, Nist A, Stiewe T, van IJcken WF, Riedlinger T, Schmitz ML, Papantonis
How cytokine-driven changes in chromatin topology are converted into gene regulatory circuits during inflammation still remains unclear. Here, we show that interleukin (IL)-1α induces acute and widespread changes in chromatin accessibility via the TAK1 kinase and NF-κB at regions that are highly enriched... |
Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture.
Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, Chan WL, Timmermann B, Wittler L, Vingron M, Mundlos S, Ibrahim DM
The genome is organized in three-dimensional units called topologically associating domains (TADs), through a process dependent on the cooperative action of cohesin and the DNA-binding factor CTCF. Genomic rearrangements of TADs have been shown to cause gene misexpression and disease, but genome-wide depletion of CT... |
Elevated cyclic-AMP represses expression of exchange protein activated by cAMP (EPAC1) by inhibiting YAP-TEAD activity and HDAC-mediated histone deacetylation.
Ebrahimighaei R, McNeill MC, Smith SA, Wray JP, Ford KL, Newby AC, Bond M
Ligand-induced activation of Exchange Protein Activated by cAMP-1 (EPAC1) is implicated in numerous physiological and pathological processes, including cardiac fibrosis where changes in EPAC1 expression have been detected. However, little is known about how EPAC1 expression is regulated. Therefore, we investigated r... |
Preformed chromatin topology assists transcriptional robustness of during limb development.
Paliou C, Guckelberger P, Schöpflin R, Heinrich V, Esposito A, Chiariello AM, Bianco S, Annunziatella C, Helmuth J, Haas S, Jerković I, Brieske N, Wittler L, Timmermann B, Nicodemi M, Vingron M, Mundlos S, Andrey G
Long-range gene regulation involves physical proximity between enhancers and promoters to generate precise patterns of gene expression in space and time. However, in some cases, proximity coincides with gene activation, whereas, in others, preformed topologies already exist before activation. In this study, we inves... |
Point mutations in the PDX1 transactivation domain impair human β-cell development and function.
Wang X, Sterr M, Ansarullah , Burtscher I, Böttcher A, Beckenbauer J, Siehler J, Meitinger T, Häring HU, Staiger H, Cernilogar FM, Schotta G, Irmler M, Beckers J, Wright CVE, Bakhti M, Lickert H
OBJECTIVE: Hundreds of missense mutations in the coding region of PDX1 exist; however, if these mutations predispose to diabetes mellitus is unknown. METHODS: In this study, we screened a large cohort of subjects with increased risk for diabetes and identified two subjects with impaired glucose tolerance carrying co... |
Global distribution of DNA hydroxymethylation and DNA methylation in chronic lymphocytic leukemia.
Wernig-Zorc S, Yadav MP, Kopparapu PK, Bemark M, Kristjansdottir HL, Andersson PO, Kanduri C, Kanduri M
BACKGROUND: Chronic lymphocytic leukemia (CLL) has been a good model system to understand the functional role of 5-methylcytosine (5-mC) in cancer progression. More recently, an oxidized form of 5-mC, 5-hydroxymethylcytosine (5-hmC) has gained lot of attention as a regulatory epigenetic modification with prognostic ... |
Mutant p63 Affects Epidermal Cell Identity through Rewiring the Enhancer Landscape.
Qu J, Tanis SEJ, Smits JPH, Kouwenhoven EN, Oti M, van den Bogaard EH, Logie C, Stunnenberg HG, van Bokhoven H, Mulder KW, Zhou H
Transcription factor p63 is a key regulator of epidermal keratinocyte proliferation and differentiation. Mutations in the p63 DNA-binding domain are associated with ectrodactyly, ectodermal dysplasia, and cleft lip/palate (EEC) syndrome. However, the underlying molecular mechanism of these mutations remains unclear.... |
Deletion of an intronic HIF-2α binding site suppresses hypoxia-induced WT1 expression.
Krueger K, Catanese L, Sciesielski LK, Kirschner KM, Scholz H
Hypoxia-inducible factors (HIFs) play a key role in the adaptation to low oxygen by interacting with hypoxia response elements (HREs) in the genome. Cellular levels of the HIF-2α transcription factor subunit influence the histopathology and clinical outcome of neuroblastoma, a malignant childhood tumor of the ... |
Sequentially acting SOX proteins orchestrate astrocyte- and oligodendrocyte-specific gene expression.
Klum S, Zaouter C, Alekseenko Z, Björklund ÅK, Hagey DW, Ericson J, Muhr J, Bergsland M
SOX transcription factors have important roles during astrocyte and oligodendrocyte development, but how glial genes are specified and activated in a sub-lineage-specific fashion remains unknown. Here, we define glial-specific gene expression in the developing spinal cord using single-cell RNA-sequencing. Moreover, ... |
Histone variant H2A.Z deposition and acetylation directs the canonical Notch signaling response.
Giaimo BD, Ferrante F, Vallejo DM, Hein K, Gutierrez-Perez I, Nist A, Stiewe T, Mittler G, Herold S, Zimmermann T, Bartkuhn M, Schwarz P, Oswald F, Dominguez M, Borggrefe T
A fundamental as yet incompletely understood feature of Notch signal transduction is a transcriptional shift from repression to activation that depends on chromatin regulation mediated by transcription factor RBP-J and associated cofactors. Incorporation of histone variants alter the functional properties of chromat... |
Epigenetic regulation of vascular NADPH oxidase expression and reactive oxygen species production by histone deacetylase-dependent mechanisms in experimental diabetes.
Manea SA, Antonescu ML, Fenyo IM, Raicu M, Simionescu M, Manea A
Reactive oxygen species (ROS) generated by up-regulated NADPH oxidase (Nox) contribute to structural-functional alterations of the vascular wall in diabetes. Epigenetic mechanisms, such as histone acetylation, emerged as important regulators of gene expression in cardiovascular disorders. Since their role in diabete... |
HMGB2 Loss upon Senescence Entry Disrupts Genomic Organization and Induces CTCF Clustering across Cell Types.
Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nürnberg P, Marenduzzo D, Rippe K, Papanton
Processes like cellular senescence are characterized by complex events giving rise to heterogeneous cell populations. However, the early molecular events driving this cascade remain elusive. We hypothesized that senescence entry is triggered by an early disruption of the cells' three-dimensional (3D) genome org... |
Genome-wide analysis of PDX1 target genes in human pancreatic progenitors.
Wang X, Sterr M, Burtscher I, Chen S, Hieronimus A, Machicao F, Staiger H, Häring HU, Lederer G, Meitinger T, Cernilogar FM, Schotta G, Irmler M, Beckers J, Hrabě de Angelis M, Ray M, Wright CVE, Bakhti M, Lickert H
OBJECTIVE: Homozygous loss-of-function mutations in the gene coding for the homeobox transcription factor (TF) PDX1 leads to pancreatic agenesis, whereas heterozygous mutations can cause Maturity-Onset Diabetes of the Young 4 (MODY4). Although the function of Pdx1 is well studied in pre-clinical models during insuli... |
A lipodystrophy-causing lamin A mutant alters conformation and epigenetic regulation of the anti-adipogenic MIR335 locus
Oldenburg A. et al.
Mutations in the Lamin A/C (LMNA) gene-encoding nuclear LMNA cause laminopathies, which include partial lipodystrophies associated with metabolic syndromes. The lipodystrophy-associated LMNA p.R482W mutation is known to impair adipogenic differentiation, but the mechanisms involved are unclear. We show in this study... |
Chromatin Immunoprecipitation (ChIP) in Mouse T-cell Lines
Giaimo B.D. et al.
Signaling pathways regulate gene expression programs via the modulation of the chromatin structure at different levels, such as by post-translational modifications (PTMs) of histone tails, the exchange of canonical histones with histone variants, and nucleosome eviction. Such regulation requires the binding of signa... |
Evolutionary re-wiring of p63 and the epigenomic regulatory landscape in keratinocytes and its potential implications on species-specific gene expression and phenotypes
Sethi I. et al.
Although epidermal keratinocyte development and differentiation proceeds in similar fashion between humans and mice, evolutionary pressures have also wrought significant species-specific physiological differences. These differences between species could arise in part, by the rewiring of regulatory network due to cha... |
Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme
Feng W. et al.
Mutations in chromatin modifier genes are frequently associated with neurodevelopmental diseases. We herein demonstrate that the chromodomain helicase DNA-binding protein 7 (Chd7), frequently associated with CHARGE syndrome, is indispensable for normal cerebellar development. Genetic inactivation of Chd7 in cerebell... |
MLL-AF9 and MLL-AF4 oncofusion proteins bind a distinct enhancer repertoire and target the RUNX1 program in 11q23 acute myeloid leukemia.
Prange KH et al.
In 11q23 leukemias, the N-terminal part of the mixed lineage leukemia (MLL) gene is fused to >60 different partner genes. In order to define a core set of MLL rearranged targets, we investigated the genome-wide binding of the MLL-AF9 and MLL-AF4 fusion proteins and associated epigenetic signatures in acute myeloi... |
Genetic variation at the 8q24.21 renal cancer susceptibility locus affects HIF binding to a MYC enhancer
Grampp S. et al.
Clear cell renal cell carcinoma (ccRCC) is characterized by loss of function of the von Hippel-Lindau tumour suppressor (VHL) and unrestrained activation of hypoxia-inducible transcription factors (HIFs). Genetic and epigenetic determinants have an impact on HIF pathways. A recent genome-wide association study on re... |
Phenotypic Plasticity through Transcriptional Regulation of the Evolutionary Hotspot Gene tan in Drosophila melanogaster
Gibert JM et al.
Phenotypic plasticity is the ability of a given genotype to produce different phenotypes in response to distinct environmental conditions. Phenotypic plasticity can be adaptive. Furthermore, it is thought to facilitate evolution. Although phenotypic plasticity is a widespread phenomenon, its molecular mechanisms are... |
Premalignant SOX2 overexpression in the fallopian tubes of ovarian cancer patients: Discovery and validation studies
Hellner K et al.
Current screening methods for ovarian cancer can only detect advanced disease. Earlier detection has proved difficult because the molecular precursors involved in the natural history of the disease are unknown. To identify early driver mutations in ovarian cancer cells, we used dense whole genome sequencing of micro... |
Genome-wide p63-regulated gene expression in differentiating epidermal keratinocytes
Otia M, Kouwenhovena EN, Zhou H
The transcription factor p63 is a key regulator in epidermal keratinocyte proliferation and differentiation. However, the role of p63 in gene regulation during these processes is not well understood. To investigate this, we recently generated genome-wide profiles of gene expression, p63 binding sites and active regu... |
Spatiotemporal control of estrogen-responsive transcription in ERα-positive breast cancer cells.
P-Y Hsu, H-K Hsu, T-H Hsiao, Z Ye, E Wang, A L Profit, I Jatoi, Y Chen, N B Kirma, V X Jin, Z D Sharp and T H-M Huang
Recruitment of transcription machinery to target promoters for aberrant gene expression has been well studied, but underlying control directed by distant-acting enhancers remains unclear in cancer development. Our previous study demonstrated that distant estrogen response elements (DEREs) located on chromosome 20q13... |
Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation
Kouwenhoven EN, Oti M, Niehues H, van Heeringen SJ, Schalkwijk J, Stunnenberg HG, van Bokhoven H, Zhou H
The transcription factor p63 plays a pivotal role in keratinocyte proliferation and differentiation in the epidermis. However, how p63 regulates epidermal genes during differentiation is not yet clear. Using epigenome profiling of differentiating human primary epidermal keratinocytes, we characterized a catalog of d... |
The Activation of IL-1-Induced Enhancers Depends on TAK1 Kinase Activity and NF-κB p65.
Jurida L, Soelch J, Bartkuhn M, Handschick K, Müller H, Newel D, Weber A, Dittrich-Breiholz O, Schneider H, Bhuju S, Saul VV, Schmitz ML, Kracht M
The inflammatory gene response requires activation of the protein kinase TAK1, but it is currently unknown how TAK1-derived signals coordinate transcriptional programs in the genome. We determined the genome-wide binding of the TAK1-controlled NF-κB subunit p65 in relation to active enhancers and promoters of transc... |
Nitric oxide-induced neuronal to glial lineage fate-change depends on NRSF/REST function in neural progenitor cells.
Bergsland M, Covacu R, Perez Estrada C, Svensson M, Brundin L
Degeneration of CNS tissue commonly occurs during neuroinflammatory conditions, such as multiple sclerosis (MS) and neurotrauma. During such conditions, neural stem/progenitor cell (NPC) populations have been suggested to provide new cells to degenerated areas. In the normal brain, NPCs from the SVZ generate neurons... |
A novel microscopy-based high-throughput screening method to identify proteins that regulate global histone modification levels.
Baas R, Lelieveld D, van Teeffelen H, Lijnzaad P, Castelijns B, van Schaik FM, Vermeulen M, Egan DA, Timmers HT, de Graaf P
Posttranslational modifications of histones play an important role in the regulation of gene expression and chromatin structure in eukaryotes. The balance between chromatin factors depositing (writers) and removing (erasers) histone marks regulates the steady-state levels of chromatin modifications. Here we describe... |