Notice (8): Undefined variable: solution_of_interest [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'product' => array(
'Product' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
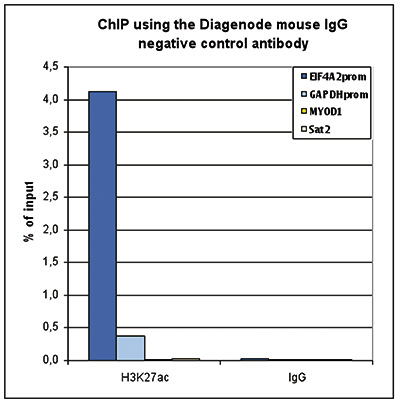
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
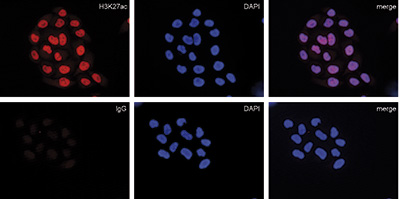
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '328',
'name' => 'Mouse IgG',
'description' => 'The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative contro',
'clonality' => '',
'isotype' => '',
'lot' => '',
'concentration' => '1.0 µg/µl',
'reactivity' => 'none',
'type' => 'Polyclonal',
'purity' => 'Protein A purified ',
'classification' => '',
'application_table' => '',
'storage_conditions' => '',
'storage_buffer' => '',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-02-24 12:10:53',
'created' => '0000-00-00 00:00:00',
'select_label' => '328 - Mouse IgG ( - 1.0 µg/µl - none - Protein A purified - Mouse)'
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/mouse-igg-15-ug-15-ul'
)
$language = 'en'
$meta_keywords = ''
$meta_description = 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.'
$meta_title = 'Mouse IgG (C15400001) | Diagenode'
$product = array(
'Product' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '328',
'name' => 'Mouse IgG',
'description' => 'The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative contro',
'clonality' => '',
'isotype' => '',
'lot' => '',
'concentration' => '1.0 µg/µl',
'reactivity' => 'none',
'type' => 'Polyclonal',
'purity' => 'Protein A purified ',
'classification' => '',
'application_table' => '',
'storage_conditions' => '',
'storage_buffer' => '',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-02-24 12:10:53',
'created' => '0000-00-00 00:00:00',
'select_label' => '328 - Mouse IgG ( - 1.0 µg/µl - none - Protein A purified - Mouse)'
),
'Slave' => array(
(int) 0 => array(
'id' => '211',
'name' => 'C15400001',
'product_id' => '1959',
'modified' => '2017-05-17 15:27:44',
'created' => '2017-05-17 15:27:44'
)
),
'Group' => array(
'Group' => array(
'id' => '211',
'name' => 'C15400001',
'product_id' => '1959',
'modified' => '2017-05-17 15:27:44',
'created' => '2017-05-17 15:27:44'
),
'Master' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '7',
'position' => '10',
'parent_id' => '1',
'name' => 'Methylated DNA immunoprecipitation',
'description' => '<div class="row extra-spaced">
<div class="small-12 medium-3 large-3 columns"><center><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank"><img src="https://www.diagenode.com/img/banners/banner-nature-publication-580.png" /></a></center></div>
<div class="small-12 medium-9 large-9 columns">
<h3>Sensitive tumour detection and classification using plasma cell-free DNA methylomes<br /><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank">Read the publication</a></h3>
<h3 class="c-article-title u-h1" data-test="article-title" itemprop="name headline">Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA<br /><a href="https://www.nature.com/articles/s41596-019-0202-2" target="_blank" title="cfMeDIP-seq Nature Method">Read the method</a></h3>
</div>
</div>
<div class="row">
<div class="large-12 columns"><span>The Methylated DNA Immunoprecipitation is based on the affinity purification of methylated and hydroxymethylated DNA using, respectively, an antibody directed against 5-methylcytosine (5-mC) in the case of MeDIP or 5-hydroxymethylcytosine (5-hmC) in the case of hMeDIP.</span><br />
<h2></h2>
<h2>How it works</h2>
<p>In brief, Methyl DNA IP is performed as follows: Genomic DNA from cultured cells or tissues is prepared, sheared, and then denatured. Then, immunoselection and immunoprecipitation can take place using the antibody directed against 5 methylcytosine and antibody binding beads. After isolation and purification is performed, the IP’d methylated DNA is ready for any subsequent analysis as qPCR, amplification, hybridization on microarrays or next generation sequencing.</p>
<h2>Applications</h2>
<div align="center"><a href="https://www.diagenode.com/en/p/magmedip-kit-x48-48-rxns" class="center alert radius button"> qPCR analysis</a></div>
<div align="center"><a href="https://www.diagenode.com/en/p/magmedip-seq-package-V2-x10" class="center alert radius button"> NGS analysis </a></div>
<h2>Advantages</h2>
<ul style="font-size: 19px;" class="nobullet">
<li><i class="fa fa-arrow-circle-right"></i> <strong>Unaffected</strong> DNA</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>High enrichment</strong> yield</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>Robust</strong> & <strong>reproducible</strong> techniques</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>NGS</strong> compatible</li>
</ul>
<h2></h2>
</div>
</div>
<div id="gtx-trans" style="position: absolute; left: 17px; top: 652.938px;">
<div class="gtx-trans-icon"></div>
</div>',
'in_footer' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'slug' => 'methylated-dna-immunoprecipitation',
'meta_keywords' => 'Methylated DNA immunoprecipitation,Epigenetic,DNA Methylation,qPCR,5 methylcytosine (5-mC)',
'meta_description' => 'Methylated DNA immunoprecipitation method is based on the affinity purification of methylated DNA using an antibody directed against 5 methylcytosine (5-mC). ',
'meta_title' => 'Methylated DNA immunoprecipitation(MeDIP) - Dna methylation | Diagenode',
'modified' => '2021-08-19 12:08:03',
'created' => '2014-09-14 05:33:34',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '22',
'position' => '40',
'parent_id' => '4',
'name' => 'Isotype controls',
'description' => '<p><span style="font-weight: 400;">Diagenode offers the negative Ctrl IgG from rabbit, rat and mouse. These extensively validated antibodies can be used as negative controls in ChIP, IF, hMeDIP or other experiments performed with specific antibodies made in rabbit, rat or mouse, respectively.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'isotype-controls',
'cookies_tag_id' => null,
'meta_keywords' => 'Isotype controls,DNA immunoprecipitation,Methylated DNA immunoprecipitation',
'meta_description' => 'Diagenode provides Isotype controls for Methylated DNA immunoprecipitation',
'meta_title' => 'Isotype controls for Methylated DNA immunoprecipitation | Diagenode',
'modified' => '2019-07-04 16:19:36',
'created' => '2014-09-30 14:23:34',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '127',
'position' => '10',
'parent_id' => '4',
'name' => 'ChIP-grade antibodies',
'description' => '<div class="row">
<div class="small-10 columns"><center></center>
<p><br />Chromatin immunoprecipitation (<b>ChIP</b>) is a technique to study the associations of proteins with the specific genomic regions in intact cells. One of the most important steps of this protocol is the immunoprecipitation of targeted protein using the antibody specifically recognizing it. The quality of antibodies used in ChIP is essential for the success of the experiment. Diagenode offers extensively validated ChIP-grade antibodies, confirmed for their specificity, and high level of performance in ChIP. Each batch is validated, and batch-specific data are available on the website.</p>
<p></p>
</div>
<div class="small-2 columns"><img src="https://www.diagenode.com/emailing/images/epi-success-guaranteed-icon.png" alt="Epigenetic success guaranteed" /></div>
</div>
<p><strong>ChIP results</strong> obtained with the antibody directed against H3K4me3 (Cat. No. <a href="../p/h3k4me3-polyclonal-antibody-premium-50-ug-50-ul">C15410003</a>). </p>
<div class="row">
<div class="small-12 medium-6 large-6 columns"><img src="https://www.diagenode.com/img/product/antibodies/C15410003-fig1-ChIP.jpg" alt="" width="400" height="315" /> </div>
<div class="small-12 medium-6 large-6 columns">
<p></p>
<p></p>
<p></p>
</div>
</div>
<p></p>
<p>Our aim at Diagenode is to offer the largest collection of highly specific <strong>ChIP-grade antibodies</strong>. We add new antibodies monthly. Find your ChIP-grade antibody in the list below and check more information about tested applications, extensive validation data, and product information.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'chip-grade-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'ChIP-grade antibodies, polyclonal antibody, monoclonal antibody, Diagenode',
'meta_description' => 'Diagenode Offers Extensively Validated ChIP-Grade Antibodies, Confirmed for their Specificity, and high level of Performance in Chromatin Immunoprecipitation ChIP',
'meta_title' => 'Chromatin immunoprecipitation ChIP-grade antibodies | Diagenode',
'modified' => '2021-07-01 10:22:38',
'created' => '2017-02-14 11:16:04',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '726',
'name' => 'Datasheet mouseIgG C15400001',
'description' => 'Datasheet description',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_mouseIgG_C15400001.pdf',
'slug' => 'datasheet-mouseigg-c15400001',
'meta_keywords' => null,
'meta_description' => null,
'modified' => '2015-07-07 11:47:44',
'created' => '2015-07-07 11:47:44',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '250',
'name' => 'product/antibodies/antibody.png',
'alt' => 'Mouse IgG',
'modified' => '2020-11-27 07:00:09',
'created' => '2015-07-17 10:12:18',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4787',
'name' => 'The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.',
'authors' => 'Garcia-Gomez E. et al.',
'description' => '<p>Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the disease (control group). In addition, eight of the PCOS patients underwent intervention with metformin (1500 mg/day) and a carbohydrate-controlled diet (type and quantity) for three months. Clinical and metabolic parameters were determined, and RT-qPCR and MeDIP-qPCR were used to evaluate gene expression and DNA methylation levels, respectively. Decreased expression levels of , , and genes and increased DNA methylation levels of the promoter were found in the endometrium of PCOS patients compared to controls. After metformin and nutritional intervention, some metabolic and clinical variables improved in PCOS patients. This intervention was associated with increased expression of , , and genes and reduced DNA methylation levels of the promoter in the endometrium of PCOS women. Our preliminary findings suggest that metformin and a carbohydrate-controlled diet improve endometrial function in PCOS patients, partly by modulating DNA methylation of the gene promoter and the expression of genes implicated in endometrial receptivity and insulin signaling.</p>',
'date' => '2023-04-01',
'pmid' => 'https://doi.org/10.3390%2Fijms24076857',
'doi' => '10.3390/ijms24076857',
'modified' => '2023-06-12 08:58:33',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4127',
'name' => 'The histone modification H3K4me3 is altered at the locus in Alzheimer'sdisease brain.',
'authors' => 'Smith, Adam et al.',
'description' => '<p>Several epigenome-wide association studies of DNA methylation have highlighted altered DNA methylation in the gene in Alzheimer's disease (AD) brain samples. However, no study has specifically examined histone modifications in the disease. We use chromatin immunoprecipitation-qPCR to quantify tri-methylation at histone 3 lysine 4 (H3K4me3) and 27 (H3K27me3) in the gene in entorhinal cortex from donors with high (n = 59) or low (n = 29) Alzheimer's disease pathology. We demonstrate decreased levels of H3K4me3, a marker of active gene transcription, with no change in H3K27me3, a marker of inactive genes. H3K4me3 is negatively correlated with DNA methylation in specific regions of the gene. Our study suggests that the gene shows altered epigenetic marks indicative of reduced gene activation in Alzheimer's disease.</p>',
'date' => '2021-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33815817',
'doi' => '10.2144/fsoa-2020-0161',
'modified' => '2021-12-07 10:16:08',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4188',
'name' => 'Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage.',
'authors' => 'Ait-Ammar A. et al.',
'description' => '<p>HIV-1 latency generates reservoirs that prevent viral eradication by the current therapies. To find strategies toward an HIV cure, detailed understandings of the molecular mechanisms underlying establishment and persistence of the reservoirs are needed. The cellular transcription factor KAP1 is known as a potent repressor of gene transcription. Here we report that KAP1 represses HIV-1 gene expression in myeloid cells including microglial cells, the major reservoir of the central nervous system. Mechanistically, KAP1 interacts and colocalizes with the viral transactivator Tat to promote its degradation via the proteasome pathway and repress HIV-1 gene expression. In myeloid models of latent HIV-1 infection, the depletion of KAP1 increased viral gene elongation and reactivated HIV-1 expression. Bound to the latent HIV-1 promoter, KAP1 associates and cooperates with CTIP2, a key epigenetic silencer of HIV-1 expression in microglial cells. In addition, Tat and CTIP2 compete for KAP1 binding suggesting a dynamic modulation of the KAP1 cellular partners upon HIV-1 infection. Altogether, our results suggest that KAP1 contributes to the establishment and the persistence of HIV-1 latency in myeloid cells.</p>',
'date' => '2021-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33514850',
'doi' => '10.1038/s41598-021-82164-w',
'modified' => '2022-01-05 15:08:41',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4082',
'name' => 'p53 directly represses human LINE1 transposons.',
'authors' => 'Tiwari, Bhavana and Jones, Amanda E and Caillet, Candace J and Das, Simantiand Royer, Stephanie K and Abrams, John M',
'description' => '<p>p53 is a potent tumor suppressor and commonly mutated in human cancers. Recently, we demonstrated that p53 genes act to restrict retrotransposons in germline tissues of flies and fish but whether this activity is conserved in somatic human cells is not known. Here we show that p53 constitutively restrains human LINE1s by cooperatively engaging sites in the 5'UTR and stimulating local deposition of repressive histone marks at these transposons. Consistent with this, the elimination of p53 or the removal of corresponding binding sites in LINE1s, prompted these retroelements to become hyperactive. Concurrently, p53 loss instigated chromosomal rearrangements linked to LINE sequences and also provoked inflammatory programs that were dependent on reverse transcriptase produced from LINE1s. Taken together, our observations establish that p53 continuously operates at the LINE1 promoter to restrict autonomous copies of these mobile elements in human cells. Our results further suggest that constitutive restriction of these retroelements may help to explain tumor suppression encoded by p53, since erupting LINE1s produced acute oncogenic threats when p53 was absent.</p>',
'date' => '2020-10-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33060137',
'doi' => '10.1101/gad.343186.120',
'modified' => '2021-03-15 16:59:03',
'created' => '2021-02-18 10:21:53',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '3953',
'name' => 'Attenuated Epigenetic Suppression of Muscle Stem Cell Necroptosis Is Required for Efficient Regeneration of Dystrophic Muscles.',
'authors' => 'Sreenivasan K, Ianni A, Künne C, Strilic B, Günther S, Perdiguero E, Krüger M, Spuler S, Offermanns S, Gómez-Del Arco P, Redondo JM, Munoz-Canoves P, Kim J, Braun T',
'description' => '<p>Somatic stem cells expand massively during tissue regeneration, which might require control of cell fitness, allowing elimination of non-competitive, potentially harmful cells. How or if such cells are removed to restore organ function is not fully understood. Here, we show that a substantial fraction of muscle stem cells (MuSCs) undergo necroptosis because of epigenetic rewiring during chronic skeletal muscle regeneration, which is required for efficient regeneration of dystrophic muscles. Inhibition of necroptosis strongly enhances suppression of MuSC expansion in a non-cell-autonomous manner. Prevention of necroptosis in MuSCs of healthy muscles is mediated by the chromatin remodeler CHD4, which directly represses the necroptotic effector Ripk3, while CHD4-dependent Ripk3 repression is dramatically attenuated in dystrophic muscles. Loss of Ripk3 repression by inactivation of Chd4 causes massive necroptosis of MuSCs, abolishing regeneration. Our study demonstrates how programmed cell death in MuSCs is tightly controlled to achieve optimal tissue regeneration.</p>',
'date' => '2020-05-19',
'pmid' => 'http://www.pubmed.gov/32433961',
'doi' => '10.1016/j.celrep.2020.107652',
'modified' => '2020-08-17 09:51:58',
'created' => '2020-08-10 12:12:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '3834',
'name' => 'Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.',
'authors' => 'Liu J, An Z, Luo J, Li J, Li F, Zhang Z',
'description' => '<p>MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of transcriptome-wide profile and potential function of m5C in splicing remains to be elucidated due to lack of isoform level m5C quantification tool. RESULTS: We developed a computational package to quantify Epitranscriptomal RNA m5C at the transcript isoform level (named Episo). Episo consists of three tools, mapper, quant and Bisulfitefq, for mapping, quantifying, and simulating RNA-BisSeq data, respectively. The high accuracy of Episo was validated using an improved m5C-specific methylated RNA immunoprecipitation (meRIP) protocol, as well as a set of in silico experiments. By applying Episo to public human and mouse RNA-BisSeq data, we found that the RNA m5C is not evenly distributed among the transcript isoforms, implying the m5C may subject to be regulated at isoform level. AVAILABILITY: Episo is released under the GNU GPLv3+ license. The resource code Episo is freely accessible from https://github.com/liujunfengtop/Episo (with Tophat/cufflink) and https://github.com/liujunfengtop/Episo/tree/master/Episo_Kallisto (with Kallisto). SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.</p>',
'date' => '2019-12-03',
'pmid' => 'http://www.pubmed.gov/31794005',
'doi' => '10.1093/bioinformatics/btz900/5651015',
'modified' => '2020-02-25 13:26:22',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '3499',
'name' => 'Centromeres License the Mitotic Condensation of Yeast Chromosome Arms.',
'authors' => 'Kruitwagen T, Chymkowitch P, Denoth-Lippuner A, Enserink J, Barral Y',
'description' => '<p>During mitosis, chromatin condensation shapes chromosomes as separate, rigid, and compact sister chromatids to facilitate their segregation. Here, we show that, unlike wild-type yeast chromosomes, non-chromosomal DNA circles and chromosomes lacking a centromere fail to condense during mitosis. The centromere promotes chromosome condensation strictly in cis through recruiting the kinases Aurora B and Bub1, which trigger the autonomous condensation of the entire chromosome. Shugoshin and the deacetylase Hst2 facilitated spreading the condensation signal to the chromosome arms. Targeting Aurora B to DNA circles or centromere-ablated chromosomes or releasing Shugoshin from PP2A-dependent inhibition bypassed the centromere requirement for condensation and enhanced the mitotic stability of DNA circles. Our data indicate that yeast cells license the chromosome-autonomous condensation of their chromatin in a centromere-dependent manner, excluding from this process non-centromeric DNA and thereby inhibiting their propagation.</p>',
'date' => '2018-10-18',
'pmid' => 'http://www.pubmed.org/30318142',
'doi' => '10.1016/j.cell.2018.09.012',
'modified' => '2019-02-27 15:44:25',
'created' => '2019-02-27 12:54:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '3235',
'name' => 'A novel FOXA1/ESR1 interacting pathway: A study of Oncomine™ breast cancer microarrays',
'authors' => 'Chaudhary S. et al.',
'description' => '<p>Forkhead box protein A1 (FOXA1) is essential for the growth and differentiation of breast epithelium, and has a favorable outcome in breast cancer (BC). Elevated <i>FOXA1</i> expression in BC also facilitates hormone responsiveness in estrogen receptor (<i>ESR</i>)-positive BC. However, the interaction between these two pathways is not fully understood. <i>FOXA1</i> and GATA binding protein 3 (<i>GATA3</i>) along with <i>ESR1</i> expression are responsible for maintaining a luminal phenotype, thus suggesting the existence of a strong association between them. The present study utilized the Oncomine™ microarray database to identify <i>FOXA1:ESR1</i> and <i>FOXA1:ESR1:GATA3</i> co-expression co-regulated genes. Oncomine™ analysis revealed 115 and 79 overlapping genes clusters in <i>FOXA1:ESR1</i> and <i>FOXA1:ESR1:GATA3</i> microarrays, respectively. Five ESR1 direct target genes [trefoil factor 1 (<i>TFF1/PS2</i>), <i>B-cell lymphoma</i> 2 (<i>BCL2</i>), seven in absentia homolog 2 (<i>SIAH2</i>), cellular myeloblastosis viral oncogene homolog (<i>CMYB</i>) and progesterone receptor (<i>PGR</i>)] were detected in the co-expression clusters. To further investigate the role of FOXA1 in ESR1-positive cells, MCF7 cells were transfected with a <i>FOXA1</i> expression plasmid, and it was observed that the direct target genes of ESR1 (<i>PS2, BCL2, SIAH2</i> and <i>PGR</i>) were significantly regulated upon transfection. Analysis of one of these target genes, <i>PS2</i>, revealed the presence of two FOXA1 binding sites in the vicinity of the estrogen response element (ERE), which was confirmed by binding assays. Under estrogen stimulation, FOXA1 protein was recruited to the FOXA1 site and could also bind to the ERE site (although in minimal amounts) in the <i>PS2</i> promoter. Co-transfection of <i>FOXA1</i>/<i>ESR1</i> expression plasmids demonstrated a significantly regulation of the target genes identified in the <i>FOXA1</i>/<i>ESR1</i> multi-arrays compared with only <i>FOXA1</i> transfection, which was suggestive of a synergistic effect of <i>ESR1</i> and <i>FOXA1</i> on the target genes. In summary, the present study identified novel <i>FOXA1</i>, <i>ESR1</i> and <i>GATA</i>3 co-expressed genes that may be involved in breast tumorigenesis.</p>',
'date' => '2017-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28789340',
'doi' => '',
'modified' => '2017-08-28 09:30:38',
'created' => '2017-08-28 09:30:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '3236',
'name' => 'TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors',
'authors' => 'Ancey P.B. et al.',
'description' => '<p>Understanding the processes that govern liver progenitor cell differentiation has important implications for the design of strategies targeting chronic liver diseases, whereby regeneration of liver tissue is critical. Although DNA methylation (5mC) and hydroxymethylation (5hmC) are highly dynamic during early embryonic development, less is known about their roles at later stages of differentiation. Using an in vitro model of hepatocyte differentiation, we show here that 5hmC precedes the expression of promoter 1 (P1)-dependent isoforms of HNF4A, a master transcription factor of hepatocyte identity. 5hmC and HNF4A expression from P1 are dependent on ten-eleven translocation (TET) dioxygenases. In turn, the liver pioneer factor FOXA2 is necessary for TET1 binding to the P1 locus. Both FOXA2 and TETs are required for the 5hmC-related switch in HNF4A expression. The epigenetic event identified here may be a key step for the establishment of the hepatocyte program by HNF4A.</p>',
'date' => '2017-07-11',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28648900',
'doi' => '',
'modified' => '2017-08-28 10:24:16',
'created' => '2017-08-28 09:44:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '3113',
'name' => 'Differentiation of Mouse Enteric Nervous System Progenitor Cells is Controlled by Endothelin 3 and Requires Regulation of Ednrb by SOX10 and ZEB2',
'authors' => 'Watanabe Y. et al.',
'description' => '<h2 class="sectionTitle"></h2>
<h3 class="sectionTitle">Background & Aims</h3>
<p>Maintenance and differentiation of progenitor cells in the developing enteric nervous system (ENS) are controlled by molecules such as the signaling protein endothelin 3 (EDN3), its receptor (the endothelin receptor type B, EDNRB), and the transcription factors SRY-box 10 (SOX10) and zinc finger E-box binding homeobox 2 (ZEB2). We used enteric progenitor cell (EPC) cultures and mice to study the roles of these proteins in enteric neurogenesis and their cross regulation.</p>
<h3 class="sectionTitle">Methods</h3>
<p>We performed studies in mice with a <em>Zeb2</em> loss-of-function mutation (<em>Zeb2</em><sup>Δ</sup>) and mice carrying a spontaneous recessive mutation that prevents conversion EDN3 to its active form (<em>Edn3</em><sup><em>ls</em></sup>). EPC cultures issued from embryos that expressed only wild-type <em>Zeb2</em> (<em>Zeb2</em><sup>+/+</sup> EPCs) or were heterozygous for the mutation (<em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs) were exposed to EDN3; we analyzed the effects on cell differentiation using immunocytochemistry. In parallel, <em>Edn3</em><sup><em>ls</em></sup> mice were crossed with <em>Zeb2</em><sup><em>Δ/+</em></sup>mice; intestinal tissues were collected from embryos for immunohistochemical analyses. We investigated regulation of the <em>EDNRB</em> gene in transactivation and chromatin immunoprecipitation assays; results were validated in functional rescue experiments using transgenes expression in EPCs from retroviral vectors.</p>
<h3 class="sectionTitle">Results</h3>
<p><em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs had increased neuronal differentiation compared to <em>Zeb2</em><sup><em>+/+</em></sup> cells. When exposed to EDN3, <em>Zeb2</em><sup>+/+</sup> EPCs continued expression of ZEB2 but did not undergo any neuronal differentiation. Incubation of <em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs with EDN3, on the other hand, resulted in only partial inhibition of neuronal differentiation. This indicated that 2 copies of <em>Zeb2</em> are required for EDN3 to prevent neuronal differentiation. Mice with combined mutations in <em>Zeb2</em> and <em>Edn3</em> (double mutants) had more severe enteric anomalies and increased neuronal differentiation compared to mice with mutations in either gene alone. The transcription factors SOX10 and ZEB2 directly activated the <em>EDNRB</em> promoter. Overexpression of EDNRB in <em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs restored inhibition of neuronal differentiation, similar to incubation of <em>Zeb2</em><sup>+/+</sup> EPCs with EDN3.</p>
<h3 class="sectionTitle">Conclusions</h3>
<p>In studies of cultured EPCs and mice, we found that control of differentiation of mouse enteric nervous system progenitor cells by EDN3 requires regulation of <em>Ednrb</em> expression by SOX10 and ZEB2.</p>',
'date' => '2017-01-05',
'pmid' => 'http://www.gastrojournal.org/article/S0016-5085(17)30002-1/abstract?referrer=http%3A%2F%2Fwww.gastrojournal.org%2Farticle%2FS0016-5085%2817%2930002-1%2Ffulltext',
'doi' => '',
'modified' => '2017-01-06 10:04:00',
'created' => '2017-01-06 10:04:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '3183',
'name' => 'High resolution methylation analysis of the HoxA5 regulatory region in different somatic tissues of laboratory mouse during development',
'authors' => 'Sinha P. et al.',
'description' => '<p>Homeobox genes encode a group of DNA binding regulatory proteins whose key function occurs in the spatial-temporal organization of genome during embryonic development and differentiation. The role of these Hox genes during ontogenesis makes it an important model for research. HoxA5 is a member of Hox gene family playing a central role during axial body patterning and morphogenesis. DNA modification studies have shown that the function of Hox genes is partly governed by the methylation-mediated gene expression regulation. Therefore the study aimed to investigate the role of epigenetic events in regulation of tissue-specific expression pattern of HoxA5 gene during mammalian development. The methodology adopted were sodium bisulfite genomic DNA sequencing, quantitative real-time PCR and chromatin-immunoprecipitation (ChIP). Methylation profiling of HoxA5 gene promoter shows higher methylation in adult as compared to fetus in various somatic tissues of mouse being highest in adult spleen. However q-PCR results show higher expression during fetal stages being highest in fetal intestine followed by brain, liver and spleen. These results clearly indicate a strict correlation between DNA methylation and tissue-specific gene expression. The findings of chromatin-immunoprecipitation (ChIP) have also reinforced that epigenetic event like DNA methylation plays important role in the regulation of tissue specific expression of HoxA5.</p>',
'date' => '2017-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28363633',
'doi' => '',
'modified' => '2017-05-22 09:48:38',
'created' => '2017-05-22 09:48:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '3054',
'name' => 'Overexpression of histone demethylase Fbxl10 leads to enhanced migration in mouse embryonic fibroblasts.',
'authors' => 'Rohde M. et al.',
'description' => '<p>Cell migration is a central process in the development and maintenance of multicellular organisms. Tissue formation during embryonic development, wound healing, immune responses and invasive tumors all require the orchestrated movement of cells to specific locations. Histone demethylase proteins alter transcription by regulating the chromatin state at specific gene loci. FBXL10 is a conserved and ubiquitously expressed member of the JmjC domain-containing histone demethylase family and is implicated in the demethylation of H3K4me3 and H3K36me2 and thereby removing active chromatin marks. However, the physiological role of FBXL10 in vivo remains largely unknown. Therefore, we established an inducible gain of function model to analyze the role of Fbxl10 and compared wild-type with Fbxl10 overexpressing mouse embryonic fibroblasts (MEFs). Our study shows that overexpression of Fbxl10 in MEFs doesn't influence the proliferation capability but leads to an enhanced migration capacity in comparison to wild-type MEFs. Transcriptome and ChIP-seq experiments demonstrated that Fbxl10 binds to genes involved in migration like Areg, Mdk, Lmnb1, Thbs1, Mgp and Cxcl12. Taken together, our results strongly suggest that Fbxl10 plays a critical role in migration by binding to the promoter region of migration-associated genes and thereby might influences cell behaviour to a possibly more aggressive phenotype.</p>',
'date' => '2016-09-17',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27646113',
'doi' => '',
'modified' => '2016-10-24 14:35:45',
'created' => '2016-10-24 14:35:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '2979',
'name' => 'Methylation of the Sox9 and Oct4 promoters and its correlation with gene expression during testicular development in the laboratory mouse',
'authors' => 'Pamnani M et al.',
'description' => '<p>Sox9 and Oct4 are two important regulatory factors involved in mammalian development. Sox9, a member of the group E Sox transcription factor family, has a crucial role in the development of the genitourinary system, while Oct4, commonly known as octamer binding transcription factor 4, belongs to class V of the transcription family. The expression of these two proteins exhibits a dynamic pattern with regard to their expression sites and levels. The aim of this study was to investigate the role of de novo methylation in the regulation of the tissue- and site-specific expression of these proteins. The dynamics of the de novo methylation of 15 CpGs and six CpGs in Sox9 and Oct4 respectively, was studied with sodium bisulfite genomic DNA sequencing in mouse testis at different developmental stages. Consistent methylation of three CpGs was observed in adult ovary in which the expression of Sox9 was feeble, while the level of methylation in somatic tissue was greater in Oct4 compared to germinal tissue. The promoter-chromatin status of Sox9 was also studied with a chromatin immune-precipitation assay.</p>',
'date' => '2016-07-04',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27381637',
'doi' => '10.1590/1678-4685-GMB-2015-0172',
'modified' => '2016-07-11 12:31:08',
'created' => '2016-07-11 12:31:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '2897',
'name' => 'Overexpression of caspase 7 is ERα dependent to affect proliferation and cell growth in breast cancer cells by targeting p21(Cip)',
'authors' => 'Chaudhary S, Madhukrishna B, Adhya AK, Keshari S, Mishra SK',
'description' => '<p>Caspase 7 (CASP7) expression has important function during cell cycle progression and cell growth in certain cancer cells and is also involved in the development and differentiation of dental tissues. However, the function of CASP7 in breast cancer cells is unclear. The aim of this study was to analyze the expression of CASP7 in breast carcinoma patients and determine the role of CASP7 in regulating tumorigenicity in breast cancer cells. In this study, we show that the CASP7 expression is high in breast carcinoma tissues compared with normal counterpart. The ectopic expression of CASP7 is significantly associated with ERα expression status and persistently elevated in different stages of the breast tumor grades. High level of CASP7 expression showed better prognosis in breast cancer patients with systemic endocrine therapy as observed from Kaplan-Meier analysis. S3 and S4, estrogen responsive element (ERE) in the CASP7 promoter, is important for estrogen-ERα-mediated CASP7 overexpression. Increased recruitment of p300, acetylated H3 and pol II in the ERE region of CASP7 promoter is observed after hormone stimulation. Ectopic expression of CASP7 in breast cancer cells results in cell growth and proliferation inhibition via p21(Cip) reduction, whereas small interfering RNA (siRNA) mediated reduction of CASP7 rescued p21(Cip) levels. We also show that pro- and active forms of CASP7 is located in the nucleus apart from cytoplasmic region of breast cancer cells. The proliferation and growth of breast cancer cells is significantly reduced by broad-spectrum peptide inhibitors and siRNA of CASP7. Taken together, our findings show that CASP7 is aberrantly expressed in breast cancer and contributes to cell growth and proliferation by downregulating p21(Cip) protein, suggesting that targeting CASP7-positive breast cancer could be one of the potential therapeutic strategies.</p>',
'date' => '2016-04-18',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27089142',
'doi' => '10.1038/oncsis.2016.12',
'modified' => '2016-04-28 10:15:00',
'created' => '2016-04-28 10:15:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '2834',
'name' => 'Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.',
'authors' => 'Rebollo R, Mager DL',
'description' => '<p>Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method used in our laboratory to detect, quantify, and compare mammalian endogenous retrovirus DNA methylation. More specifically we describe methylated DNA immunoprecipitation (MeDIP) followed by quantitative PCR.</p>',
'date' => '2016-02-20',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26895065',
'doi' => '10.1007/978-1-4939-3372-3_23',
'modified' => '2016-03-02 10:37:15',
'created' => '2016-03-02 10:22:16',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '2842',
'name' => 'Chromatin Immunoprecipitation Assay for the Identification of Arabidopsis Protein-DNA Interactions In Vivo',
'authors' => 'Komar DN, Mouriz A, Jarillo JA, Piñeiro M',
'description' => '<p>Intricate gene regulatory networks orchestrate biological processes and developmental transitions in plants. Selective transcriptional activation and silencing of genes mediate the response of plants to environmental signals and developmental cues. Therefore, insights into the mechanisms that control plant gene expression are essential to gain a deep understanding of how biological processes are regulated in plants. The chromatin immunoprecipitation (ChIP) technique described here is a procedure to identify the DNA-binding sites of proteins in genes or genomic regions of the model species Arabidopsis thaliana. The interactions with DNA of proteins of interest such as transcription factors, chromatin proteins or posttranslationally modified versions of histones can be efficiently analyzed with the ChIP protocol. This method is based on the fixation of protein-DNA interactions in vivo, random fragmentation of chromatin, immunoprecipitation of protein-DNA complexes with specific antibodies, and quantification of the DNA associated with the protein of interest by PCR techniques. The use of this methodology in Arabidopsis has contributed significantly to unveil transcriptional regulatory mechanisms that control a variety of plant biological processes. This approach allowed the identification of the binding sites of the Arabidopsis chromatin protein EBS to regulatory regions of the master gene of flowering FT. The impact of this protein in the accumulation of particular histone marks in the genomic region of FT was also revealed through ChIP analysis.</p>',
'date' => '2016-01-14',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26863263',
'doi' => '10.3791/53422',
'modified' => '2017-01-04 14:16:52',
'created' => '2016-03-09 17:05:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '2801',
'name' => 'The genetic association of RUNX3 with ankylosing spondylitis can be explained by allele-specific effects on IRF4 recruitment that alter gene expression',
'authors' => 'Matteo Vecellio, Amity R Roberts, Carla J Cohen, Adrian Cortes, Julian C Knight, Paul Bowness, B Paul Wordsworth',
'description' => '<p>The authors sought to identify the functional basis for the genetic association of single nucleotide polymorphisms (SNP), upstream of the RUNX3 promoter, with ankylosing spondylitis (AS). They performed conditional analysis of genetic association data and used ENCODE data on chromatin remodelling and transcription factor (TF) binding sites to identify the primary AS-associated regulatory SNP in the RUNX3 region. The functional effects of this SNP were tested in luciferase reporter assays. Its effects on TF binding were investigated by electrophoretic mobility gel shift assays and chromatin immunoprecipitation. RUNX3 mRNA levels were compared in primary CD8+ T cells of AS risk and protective genotypes by real-time PCR. They identified a regulatory region upstream of RUNX3 that is modulated byrs4648889. The risk allele decreases TF binding (including IRF4) and reduces reporter activity and RUNX3 expression. These findings may have important implications for understanding the role of T cells and other immune cells in AS.</p>',
'date' => '2015-10-09',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26452539',
'doi' => '10.1136/annrheumdis-2015-207490',
'modified' => '2016-04-25 09:56:04',
'created' => '2015-12-07 06:37:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '2852',
'name' => 'ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells',
'authors' => 'Al-Furoukh N, Ianni A, Nolte H, Hölper S, Krüger M, Wanrooij S, Braun T',
'description' => '<p>Proteostasis is crucial for life and maintained by cellular chaperones and proteases. One major mitochondrial protease is the ClpXP complex, which is comprised of a catalytic ClpX subunit and a proteolytic ClpP subunit. Based on two separate observations, we hypothesized that ClpX may play a leading role in the cellular function of ClpXP. Therefore, we analyzed the effect of ClpX overexpression on a myoblast proteome by quantitative proteomics. ClpX overexpression results in the upregulation of markers of the mitochondrial proteostasis pathway, known as the "mitochondrial unfolded protein response" (UPRmt). Although this pathway is described in detail in Caenorhabditis elegans, it is not clear whether it is conserved in mammals. Therefore, we compared features of the classical nematode UPRmt with our mammalian ClpX-triggered UPRmt dataset. We show that they share the same retrograde mitochondria-to-nucleus signaling pathway that involves the key UPRmt transcription factor CHOP (also known as Ddit3, CEBPZ or GADD153). In conclusion, our data confirm the existence of a mammalian UPRmt that has great similarity to the C. elegans pathway. Furthermore, our results illustrate that ClpX overexpression is a good and simple model to study the underlying mechanisms of the UPRmt in mammalian cells.</p>',
'date' => '2015-10-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26142927',
'doi' => '10.1016/j.bbamcr.2015.06.016',
'modified' => '2016-03-14 10:20:09',
'created' => '2016-03-14 10:20:09',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '2890',
'name' => 'Endothelial Msx1 transduces hemodynamic changes into an arteriogenic remodeling response.',
'authors' => 'Vandersmissen I, Craps S, Depypere M, Coppiello G, van Gastel N, Maes F, Carmeliet G, Schrooten J, Jones EA, Umans L, Devlieger R, Koole M, Gheysens O, Zwijsen A, Aranguren XL, Luttun A',
'description' => '<p>Collateral remodeling is critical for blood flow restoration in peripheral arterial disease and is triggered by increasing fluid shear stress in preexisting collateral arteries. So far, no arterial-specific mediators of this mechanotransduction response have been identified. We show that muscle segment homeobox 1 (MSX1) acts exclusively in collateral arterial endothelium to transduce the extrinsic shear stimulus into an arteriogenic remodeling response. MSX1 was specifically up-regulated in remodeling collateral arteries. MSX1 induction in collateral endothelial cells (ECs) was shear stress driven and downstream of canonical bone morphogenetic protein-SMAD signaling. Flow recovery and collateral remodeling were significantly blunted in EC-specific Msx1/2 knockout mice. Mechanistically, MSX1 linked the arterial shear stimulus to arteriogenic remodeling by activating the endothelial but not medial layer to a proinflammatory state because EC but not smooth muscle cellMsx1/2 knockout mice had reduced leukocyte recruitment to remodeling collateral arteries. This reduced leukocyte infiltration in EC Msx1/2 knockout mice originated from decreased levels of intercellular adhesion molecule 1 (ICAM1)/vascular cell adhesion molecule 1 (VCAM1), whose expression was also in vitro driven by promoter binding of MSX1.</p>',
'date' => '2015-09-28',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26391659',
'doi' => ' 10.1083/jcb.201502003',
'modified' => '2016-04-12 10:44:22',
'created' => '2016-04-12 10:44:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '2846',
'name' => 'DNA methylation directs functional maturation of pancreatic β cells',
'authors' => 'Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matveyenko A, Bhushan A.',
'description' => '<p>Pancreatic β cells secrete insulin in response to postprandial increases in glucose levels to prevent hyperglycemia and inhibit insulin secretion under fasting conditions to protect against hypoglycemia. β cells lack this functional capability at birth and acquire glucose-stimulated insulin secretion (GSIS) during neonatal life. Here, we have shown that during postnatal life, the de novo DNA methyltransferase DNMT3A initiates a metabolic program by repressing key genes, thereby enabling the coupling of insulin secretion to glucose levels. In a murine model, β cell-specific deletion of Dnmt3a prevented the metabolic switch, resulting in loss of GSIS. DNMT3A bound to the promoters of the genes encoding hexokinase 1 (HK1) and lactate dehydrogenase A (LDHA) - both of which regulate the metabolic switch - and knockdown of these two key DNMT3A targets restored the GSIS response in islets from animals with β cell-specific Dnmt3a deletion. Furthermore, DNA methylation-mediated repression of glucose-secretion decoupling genes to modulate GSIS was conserved in human β cells. Together, our results reveal a role for DNA methylation to direct the acquisition of pancreatic β cell function.</p>',
'date' => '2015-07-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26098213',
'doi' => '10.1172/JCI79956',
'modified' => '2016-03-11 14:16:44',
'created' => '2016-03-11 14:16:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '2049',
'name' => 'Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions.',
'authors' => 'Tapias A, Zhou ZW, Shi Y, Chong Z, Wang P, Groth M, Platzer M, Huttner W, Herceg Z, Yang YG, Wang ZQ',
'description' => 'Fate decisions in neural progenitor cells are orchestrated via multiple pathways, and the role of histone acetylation in these decisions has been ascribed to a general function promoting gene activation. Here, we show that the histone acetyltransferase (HAT) cofactor transformation/transcription domain-associated protein (Trrap) specifically regulates activation of cell-cycle genes, thereby integrating discrete cell-intrinsic programs of cell-cycle progression and epigenetic regulation of gene transcription in order to control neurogenesis. Deletion of Trrap impairs recruitment of HATs and transcriptional machinery specifically to E2F cell-cycle target genes, disrupting their transcription with consequent cell-cycle lengthening specifically within cortical apical neural progenitors (APs). Consistently, Trrap conditional mutants exhibit microcephaly because of premature differentiation of APs into intermediate basal progenitors and neurons, and overexpressing cell-cycle regulators in vivo can rescue these premature differentiation defects. These results demonstrate an essential and highly specific role for Trrap-mediated histone regulation in controlling cell-cycle progression and neurogenesis.',
'date' => '2014-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24792116',
'doi' => '',
'modified' => '2015-07-24 15:39:02',
'created' => '2015-07-24 15:39:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '1493',
'name' => 'Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks.',
'authors' => 'Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gäbel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D',
'description' => 'The chromosome 9p21 (Chr9p21) locus of coronary artery disease has been identified in the first surge of genome-wide association and is the strongest genetic factor of atherosclerosis known today. Chr9p21 encodes the long non-coding RNA (ncRNA) antisense non-coding RNA in the INK4 locus (ANRIL). ANRIL expression is associated with the Chr9p21 genotype and correlated with atherosclerosis severity. Here, we report on the molecular mechanisms through which ANRIL regulates target-genes in trans, leading to increased cell proliferation, increased cell adhesion and decreased apoptosis, which are all essential mechanisms of atherogenesis. Importantly, trans-regulation was dependent on Alu motifs, which marked the promoters of ANRIL target genes and were mirrored in ANRIL RNA transcripts. ANRIL bound Polycomb group proteins that were highly enriched in the proximity of Alu motifs across the genome and were recruited to promoters of target genes upon ANRIL over-expression. The functional relevance of Alu motifs in ANRIL was confirmed by deletion and mutagenesis, reversing trans-regulation and atherogenic cell functions. ANRIL-regulated networks were confirmed in 2280 individuals with and without coronary artery disease and functionally validated in primary cells from patients carrying the Chr9p21 risk allele. Our study provides a molecular mechanism for pro-atherogenic effects of ANRIL at Chr9p21 and suggests a novel role for Alu elements in epigenetic gene regulation by long ncRNAs.',
'date' => '2013-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23861667',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '1285',
'name' => 'Genome-wide analysis of LXRα activation reveals new transcriptional networks in human atherosclerotic foam cells.',
'authors' => 'Feldmann R, Fischer C, Kodelja V, Behrens S, Haas S, Vingron M, Timmermann B, Geikowski A, Sauer S',
'description' => 'Increased physiological levels of oxysterols are major risk factors for developing atherosclerosis and cardiovascular disease. Lipid-loaded macrophages, termed foam cells, are important during the early development of atherosclerotic plaques. To pursue the hypothesis that ligand-based modulation of the nuclear receptor LXRα is crucial for cell homeostasis during atherosclerotic processes, we analysed genome-wide the action of LXRα in foam cells and macrophages. By integrating chromatin immunoprecipitation-sequencing (ChIP-seq) and gene expression profile analyses, we generated a highly stringent set of 186 LXRα target genes. Treatment with the nanomolar-binding ligand T0901317 and subsequent auto-regulatory LXRα activation resulted in sequence-dependent sharpening of the genome-binding patterns of LXRα. LXRα-binding loci that correlated with differential gene expression revealed 32 novel target genes with potential beneficial effects, which in part explained the implications of disease-associated genetic variation data. These observations identified highly integrated LXRα ligand-dependent transcriptional networks, including the APOE/C1/C4/C2-gene cluster, which contribute to the reversal of cholesterol efflux and the dampening of inflammation processes in foam cells to prevent atherogenesis.',
'date' => '2013-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23393188',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '1078',
'name' => 'New partners in regulation of gene expression: the enhancer of trithorax and polycomb corto interacts with methylated ribosomal protein l12 via its chromodomain.',
'authors' => 'Coléno-Costes A, Jang SM, de Vanssay A, Rougeot J, Bouceba T, Randsholt NB, Gibert JM, Le Crom S, Mouchel-Vielh E, Bloyer S, Peronnet F',
'description' => 'Chromodomains are found in many regulators of chromatin structure, and most of them recognize methylated lysines on histones. Here, we investigate the role of the Drosophila melanogaster protein Corto's chromodomain. The Enhancer of Trithorax and Polycomb Corto is involved in both silencing and activation of gene expression. Over-expression of the Corto chromodomain (CortoCD) in transgenic flies shows that it is a chromatin-targeting module, critical for Corto function. Unexpectedly, mass spectrometry analysis reveals that polypeptides pulled down by CortoCD from nuclear extracts correspond to ribosomal proteins. Furthermore, real-time interaction analyses demonstrate that CortoCD binds with high affinity RPL12 tri-methylated on lysine 3. Corto and RPL12 co-localize with active epigenetic marks on polytene chromosomes, suggesting that both are involved in fine-tuning transcription of genes in open chromatin. RNA-seq based transcriptomes of wing imaginal discs over-expressing either CortoCD or RPL12 reveal that both factors deregulate large sets of common genes, which are enriched in heat-response and ribosomal protein genes, suggesting that they could be implicated in dynamic coordination of ribosome biogenesis. Chromatin immunoprecipitation experiments show that Corto and RPL12 bind hsp70 and are similarly recruited on gene body after heat shock. Hence, Corto and RPL12 could be involved together in regulation of gene transcription. We discuss whether pseudo-ribosomal complexes composed of various ribosomal proteins might participate in regulation of gene expression in connection with chromatin regulators.',
'date' => '2012-10-11',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23071455',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '930',
'name' => 'The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts',
'authors' => 'Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J',
'description' => 'Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain and a Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts (MEFs) stably over-expressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the non-coding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2 and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches, showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression and the metabolic control of fibroblasts. ',
'date' => '2012-07-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22825849',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '352',
'name' => 'Mouse IgG SDS US en',
'language' => 'en',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-US-en-GHS_1_0.pdf',
'countries' => 'US',
'modified' => '2020-06-09 15:24:43',
'created' => '2020-06-09 15:24:43',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '350',
'name' => 'Mouse IgG SDS GB en',
'language' => 'en',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-GB-en-GHS_1_0.pdf',
'countries' => 'GB',
'modified' => '2020-06-09 15:23:14',
'created' => '2020-06-09 15:23:14',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '348',
'name' => 'Mouse IgG SDS ES es',
'language' => 'es',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-ES-es-GHS_1_0.pdf',
'countries' => 'ES',
'modified' => '2020-06-09 15:22:03',
'created' => '2020-06-09 15:22:03',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '347',
'name' => 'Mouse IgG SDS DE de',
'language' => 'de',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-DE-de-GHS_1_0.pdf',
'countries' => 'DE',
'modified' => '2020-06-09 15:21:25',
'created' => '2020-06-09 15:21:25',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '351',
'name' => 'Mouse IgG SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-JP-ja-GHS_1_0.pdf',
'countries' => 'JP',
'modified' => '2020-06-09 15:23:45',
'created' => '2020-06-09 15:23:45',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '346',
'name' => 'Mouse IgG SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-BE-nl-GHS_1_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-09 15:20:49',
'created' => '2020-06-09 15:20:49',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '345',
'name' => 'Mouse IgG SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-BE-fr-GHS_1_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-09 15:19:47',
'created' => '2020-06-09 15:19:47',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '349',
'name' => 'Mouse IgG SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-FR-fr-GHS_1_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-09 15:22:38',
'created' => '2020-06-09 15:22:38',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/mouse-igg-15-ug-15-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
)
$pro = array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/bioruptor-pico-sonication-device"><img src="/img/product/shearing_technologies/bioruptor_pico.jpg" alt="Bioruptor pico next gen sequencing " class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">B01060010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Bioruptor® Pico sonication device</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '1862',
'product_id' => '1959',
'related_id' => '1787'
),
'Image' => array(
(int) 0 => array(
'id' => '133',
'name' => 'product/shearing_technologies/bioruptor_pico.jpg',
'alt' => 'Bioruptor pico next gen sequencing ',
'modified' => '2020-11-27 10:00:30',
'created' => '2015-06-10 17:28:55',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1772',
'name' => 'product/shearing_technologies/B010600010.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 15:41:46',
'created' => '2018-02-14 15:41:46',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1773',
'name' => 'product/shearing_technologies/B010600010-2.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 16:18:03',
'created' => '2018-02-14 15:42:00',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1774',
'name' => 'product/shearing_technologies/B01060010-bioruptor-pico-cooler.jpg',
'alt' => 'Pico + cooler',
'modified' => '2018-03-06 11:01:33',
'created' => '2018-03-06 11:01:33',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
'id' => '4167',
'product_id' => '1959',
'application_id' => '43'
)
)
$slugs = array(
(int) 0 => 'chip-qpcr-antibodies'
)
$applications = array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'locale' => 'eng'
)
$description = ''
$name = 'ChIP-qPCR (ab)'
$document = array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '2068',
'product_id' => '1959',
'document_id' => '38'
)
)
$sds = array(
'id' => '349',
'name' => 'Mouse IgG SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-FR-fr-GHS_1_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-09 15:22:38',
'created' => '2020-06-09 15:22:38',
'ProductsSafetySheet' => array(
'id' => '681',
'product_id' => '1959',
'safety_sheet_id' => '349'
)
)
$publication = array(
'id' => '930',
'name' => 'The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts',
'authors' => 'Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J',
'description' => 'Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain and a Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts (MEFs) stably over-expressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the non-coding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2 and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches, showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression and the metabolic control of fibroblasts. ',
'date' => '2012-07-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22825849',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
'id' => '1075',
'product_id' => '1959',
'publication_id' => '930'
)
)
$externalLink = ' <a href="http://www.ncbi.nlm.nih.gov/pubmed/22825849" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: header [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'product' => array(
'Product' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '328',
'name' => 'Mouse IgG',
'description' => 'The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative contro',
'clonality' => '',
'isotype' => '',
'lot' => '',
'concentration' => '1.0 µg/µl',
'reactivity' => 'none',
'type' => 'Polyclonal',
'purity' => 'Protein A purified ',
'classification' => '',
'application_table' => '',
'storage_conditions' => '',
'storage_buffer' => '',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-02-24 12:10:53',
'created' => '0000-00-00 00:00:00',
'select_label' => '328 - Mouse IgG ( - 1.0 µg/µl - none - Protein A purified - Mouse)'
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/mouse-igg-15-ug-15-ul'
)
$language = 'en'
$meta_keywords = ''
$meta_description = 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.'
$meta_title = 'Mouse IgG (C15400001) | Diagenode'
$product = array(
'Product' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '328',
'name' => 'Mouse IgG',
'description' => 'The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative contro',
'clonality' => '',
'isotype' => '',
'lot' => '',
'concentration' => '1.0 µg/µl',
'reactivity' => 'none',
'type' => 'Polyclonal',
'purity' => 'Protein A purified ',
'classification' => '',
'application_table' => '',
'storage_conditions' => '',
'storage_buffer' => '',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-02-24 12:10:53',
'created' => '0000-00-00 00:00:00',
'select_label' => '328 - Mouse IgG ( - 1.0 µg/µl - none - Protein A purified - Mouse)'
),
'Slave' => array(
(int) 0 => array(
'id' => '211',
'name' => 'C15400001',
'product_id' => '1959',
'modified' => '2017-05-17 15:27:44',
'created' => '2017-05-17 15:27:44'
)
),
'Group' => array(
'Group' => array(
'id' => '211',
'name' => 'C15400001',
'product_id' => '1959',
'modified' => '2017-05-17 15:27:44',
'created' => '2017-05-17 15:27:44'
),
'Master' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '7',
'position' => '10',
'parent_id' => '1',
'name' => 'Methylated DNA immunoprecipitation',
'description' => '<div class="row extra-spaced">
<div class="small-12 medium-3 large-3 columns"><center><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank"><img src="https://www.diagenode.com/img/banners/banner-nature-publication-580.png" /></a></center></div>
<div class="small-12 medium-9 large-9 columns">
<h3>Sensitive tumour detection and classification using plasma cell-free DNA methylomes<br /><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank">Read the publication</a></h3>
<h3 class="c-article-title u-h1" data-test="article-title" itemprop="name headline">Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA<br /><a href="https://www.nature.com/articles/s41596-019-0202-2" target="_blank" title="cfMeDIP-seq Nature Method">Read the method</a></h3>
</div>
</div>
<div class="row">
<div class="large-12 columns"><span>The Methylated DNA Immunoprecipitation is based on the affinity purification of methylated and hydroxymethylated DNA using, respectively, an antibody directed against 5-methylcytosine (5-mC) in the case of MeDIP or 5-hydroxymethylcytosine (5-hmC) in the case of hMeDIP.</span><br />
<h2></h2>
<h2>How it works</h2>
<p>In brief, Methyl DNA IP is performed as follows: Genomic DNA from cultured cells or tissues is prepared, sheared, and then denatured. Then, immunoselection and immunoprecipitation can take place using the antibody directed against 5 methylcytosine and antibody binding beads. After isolation and purification is performed, the IP’d methylated DNA is ready for any subsequent analysis as qPCR, amplification, hybridization on microarrays or next generation sequencing.</p>
<h2>Applications</h2>
<div align="center"><a href="https://www.diagenode.com/en/p/magmedip-kit-x48-48-rxns" class="center alert radius button"> qPCR analysis</a></div>
<div align="center"><a href="https://www.diagenode.com/en/p/magmedip-seq-package-V2-x10" class="center alert radius button"> NGS analysis </a></div>
<h2>Advantages</h2>
<ul style="font-size: 19px;" class="nobullet">
<li><i class="fa fa-arrow-circle-right"></i> <strong>Unaffected</strong> DNA</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>High enrichment</strong> yield</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>Robust</strong> & <strong>reproducible</strong> techniques</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>NGS</strong> compatible</li>
</ul>
<h2></h2>
</div>
</div>
<div id="gtx-trans" style="position: absolute; left: 17px; top: 652.938px;">
<div class="gtx-trans-icon"></div>
</div>',
'in_footer' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'slug' => 'methylated-dna-immunoprecipitation',
'meta_keywords' => 'Methylated DNA immunoprecipitation,Epigenetic,DNA Methylation,qPCR,5 methylcytosine (5-mC)',
'meta_description' => 'Methylated DNA immunoprecipitation method is based on the affinity purification of methylated DNA using an antibody directed against 5 methylcytosine (5-mC). ',
'meta_title' => 'Methylated DNA immunoprecipitation(MeDIP) - Dna methylation | Diagenode',
'modified' => '2021-08-19 12:08:03',
'created' => '2014-09-14 05:33:34',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '22',
'position' => '40',
'parent_id' => '4',
'name' => 'Isotype controls',
'description' => '<p><span style="font-weight: 400;">Diagenode offers the negative Ctrl IgG from rabbit, rat and mouse. These extensively validated antibodies can be used as negative controls in ChIP, IF, hMeDIP or other experiments performed with specific antibodies made in rabbit, rat or mouse, respectively.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'isotype-controls',
'cookies_tag_id' => null,
'meta_keywords' => 'Isotype controls,DNA immunoprecipitation,Methylated DNA immunoprecipitation',
'meta_description' => 'Diagenode provides Isotype controls for Methylated DNA immunoprecipitation',
'meta_title' => 'Isotype controls for Methylated DNA immunoprecipitation | Diagenode',
'modified' => '2019-07-04 16:19:36',
'created' => '2014-09-30 14:23:34',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '127',
'position' => '10',
'parent_id' => '4',
'name' => 'ChIP-grade antibodies',
'description' => '<div class="row">
<div class="small-10 columns"><center></center>
<p><br />Chromatin immunoprecipitation (<b>ChIP</b>) is a technique to study the associations of proteins with the specific genomic regions in intact cells. One of the most important steps of this protocol is the immunoprecipitation of targeted protein using the antibody specifically recognizing it. The quality of antibodies used in ChIP is essential for the success of the experiment. Diagenode offers extensively validated ChIP-grade antibodies, confirmed for their specificity, and high level of performance in ChIP. Each batch is validated, and batch-specific data are available on the website.</p>
<p></p>
</div>
<div class="small-2 columns"><img src="https://www.diagenode.com/emailing/images/epi-success-guaranteed-icon.png" alt="Epigenetic success guaranteed" /></div>
</div>
<p><strong>ChIP results</strong> obtained with the antibody directed against H3K4me3 (Cat. No. <a href="../p/h3k4me3-polyclonal-antibody-premium-50-ug-50-ul">C15410003</a>). </p>
<div class="row">
<div class="small-12 medium-6 large-6 columns"><img src="https://www.diagenode.com/img/product/antibodies/C15410003-fig1-ChIP.jpg" alt="" width="400" height="315" /> </div>
<div class="small-12 medium-6 large-6 columns">
<p></p>
<p></p>
<p></p>
</div>
</div>
<p></p>
<p>Our aim at Diagenode is to offer the largest collection of highly specific <strong>ChIP-grade antibodies</strong>. We add new antibodies monthly. Find your ChIP-grade antibody in the list below and check more information about tested applications, extensive validation data, and product information.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'chip-grade-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'ChIP-grade antibodies, polyclonal antibody, monoclonal antibody, Diagenode',
'meta_description' => 'Diagenode Offers Extensively Validated ChIP-Grade Antibodies, Confirmed for their Specificity, and high level of Performance in Chromatin Immunoprecipitation ChIP',
'meta_title' => 'Chromatin immunoprecipitation ChIP-grade antibodies | Diagenode',
'modified' => '2021-07-01 10:22:38',
'created' => '2017-02-14 11:16:04',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '726',
'name' => 'Datasheet mouseIgG C15400001',
'description' => 'Datasheet description',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_mouseIgG_C15400001.pdf',
'slug' => 'datasheet-mouseigg-c15400001',
'meta_keywords' => null,
'meta_description' => null,
'modified' => '2015-07-07 11:47:44',
'created' => '2015-07-07 11:47:44',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '250',
'name' => 'product/antibodies/antibody.png',
'alt' => 'Mouse IgG',
'modified' => '2020-11-27 07:00:09',
'created' => '2015-07-17 10:12:18',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4787',
'name' => 'The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.',
'authors' => 'Garcia-Gomez E. et al.',
'description' => '<p>Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the disease (control group). In addition, eight of the PCOS patients underwent intervention with metformin (1500 mg/day) and a carbohydrate-controlled diet (type and quantity) for three months. Clinical and metabolic parameters were determined, and RT-qPCR and MeDIP-qPCR were used to evaluate gene expression and DNA methylation levels, respectively. Decreased expression levels of , , and genes and increased DNA methylation levels of the promoter were found in the endometrium of PCOS patients compared to controls. After metformin and nutritional intervention, some metabolic and clinical variables improved in PCOS patients. This intervention was associated with increased expression of , , and genes and reduced DNA methylation levels of the promoter in the endometrium of PCOS women. Our preliminary findings suggest that metformin and a carbohydrate-controlled diet improve endometrial function in PCOS patients, partly by modulating DNA methylation of the gene promoter and the expression of genes implicated in endometrial receptivity and insulin signaling.</p>',
'date' => '2023-04-01',
'pmid' => 'https://doi.org/10.3390%2Fijms24076857',
'doi' => '10.3390/ijms24076857',
'modified' => '2023-06-12 08:58:33',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4127',
'name' => 'The histone modification H3K4me3 is altered at the locus in Alzheimer'sdisease brain.',
'authors' => 'Smith, Adam et al.',
'description' => '<p>Several epigenome-wide association studies of DNA methylation have highlighted altered DNA methylation in the gene in Alzheimer's disease (AD) brain samples. However, no study has specifically examined histone modifications in the disease. We use chromatin immunoprecipitation-qPCR to quantify tri-methylation at histone 3 lysine 4 (H3K4me3) and 27 (H3K27me3) in the gene in entorhinal cortex from donors with high (n = 59) or low (n = 29) Alzheimer's disease pathology. We demonstrate decreased levels of H3K4me3, a marker of active gene transcription, with no change in H3K27me3, a marker of inactive genes. H3K4me3 is negatively correlated with DNA methylation in specific regions of the gene. Our study suggests that the gene shows altered epigenetic marks indicative of reduced gene activation in Alzheimer's disease.</p>',
'date' => '2021-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33815817',
'doi' => '10.2144/fsoa-2020-0161',
'modified' => '2021-12-07 10:16:08',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4188',
'name' => 'Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage.',
'authors' => 'Ait-Ammar A. et al.',
'description' => '<p>HIV-1 latency generates reservoirs that prevent viral eradication by the current therapies. To find strategies toward an HIV cure, detailed understandings of the molecular mechanisms underlying establishment and persistence of the reservoirs are needed. The cellular transcription factor KAP1 is known as a potent repressor of gene transcription. Here we report that KAP1 represses HIV-1 gene expression in myeloid cells including microglial cells, the major reservoir of the central nervous system. Mechanistically, KAP1 interacts and colocalizes with the viral transactivator Tat to promote its degradation via the proteasome pathway and repress HIV-1 gene expression. In myeloid models of latent HIV-1 infection, the depletion of KAP1 increased viral gene elongation and reactivated HIV-1 expression. Bound to the latent HIV-1 promoter, KAP1 associates and cooperates with CTIP2, a key epigenetic silencer of HIV-1 expression in microglial cells. In addition, Tat and CTIP2 compete for KAP1 binding suggesting a dynamic modulation of the KAP1 cellular partners upon HIV-1 infection. Altogether, our results suggest that KAP1 contributes to the establishment and the persistence of HIV-1 latency in myeloid cells.</p>',
'date' => '2021-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33514850',
'doi' => '10.1038/s41598-021-82164-w',
'modified' => '2022-01-05 15:08:41',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4082',
'name' => 'p53 directly represses human LINE1 transposons.',
'authors' => 'Tiwari, Bhavana and Jones, Amanda E and Caillet, Candace J and Das, Simantiand Royer, Stephanie K and Abrams, John M',
'description' => '<p>p53 is a potent tumor suppressor and commonly mutated in human cancers. Recently, we demonstrated that p53 genes act to restrict retrotransposons in germline tissues of flies and fish but whether this activity is conserved in somatic human cells is not known. Here we show that p53 constitutively restrains human LINE1s by cooperatively engaging sites in the 5'UTR and stimulating local deposition of repressive histone marks at these transposons. Consistent with this, the elimination of p53 or the removal of corresponding binding sites in LINE1s, prompted these retroelements to become hyperactive. Concurrently, p53 loss instigated chromosomal rearrangements linked to LINE sequences and also provoked inflammatory programs that were dependent on reverse transcriptase produced from LINE1s. Taken together, our observations establish that p53 continuously operates at the LINE1 promoter to restrict autonomous copies of these mobile elements in human cells. Our results further suggest that constitutive restriction of these retroelements may help to explain tumor suppression encoded by p53, since erupting LINE1s produced acute oncogenic threats when p53 was absent.</p>',
'date' => '2020-10-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33060137',
'doi' => '10.1101/gad.343186.120',
'modified' => '2021-03-15 16:59:03',
'created' => '2021-02-18 10:21:53',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '3953',
'name' => 'Attenuated Epigenetic Suppression of Muscle Stem Cell Necroptosis Is Required for Efficient Regeneration of Dystrophic Muscles.',
'authors' => 'Sreenivasan K, Ianni A, Künne C, Strilic B, Günther S, Perdiguero E, Krüger M, Spuler S, Offermanns S, Gómez-Del Arco P, Redondo JM, Munoz-Canoves P, Kim J, Braun T',
'description' => '<p>Somatic stem cells expand massively during tissue regeneration, which might require control of cell fitness, allowing elimination of non-competitive, potentially harmful cells. How or if such cells are removed to restore organ function is not fully understood. Here, we show that a substantial fraction of muscle stem cells (MuSCs) undergo necroptosis because of epigenetic rewiring during chronic skeletal muscle regeneration, which is required for efficient regeneration of dystrophic muscles. Inhibition of necroptosis strongly enhances suppression of MuSC expansion in a non-cell-autonomous manner. Prevention of necroptosis in MuSCs of healthy muscles is mediated by the chromatin remodeler CHD4, which directly represses the necroptotic effector Ripk3, while CHD4-dependent Ripk3 repression is dramatically attenuated in dystrophic muscles. Loss of Ripk3 repression by inactivation of Chd4 causes massive necroptosis of MuSCs, abolishing regeneration. Our study demonstrates how programmed cell death in MuSCs is tightly controlled to achieve optimal tissue regeneration.</p>',
'date' => '2020-05-19',
'pmid' => 'http://www.pubmed.gov/32433961',
'doi' => '10.1016/j.celrep.2020.107652',
'modified' => '2020-08-17 09:51:58',
'created' => '2020-08-10 12:12:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '3834',
'name' => 'Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.',
'authors' => 'Liu J, An Z, Luo J, Li J, Li F, Zhang Z',
'description' => '<p>MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of transcriptome-wide profile and potential function of m5C in splicing remains to be elucidated due to lack of isoform level m5C quantification tool. RESULTS: We developed a computational package to quantify Epitranscriptomal RNA m5C at the transcript isoform level (named Episo). Episo consists of three tools, mapper, quant and Bisulfitefq, for mapping, quantifying, and simulating RNA-BisSeq data, respectively. The high accuracy of Episo was validated using an improved m5C-specific methylated RNA immunoprecipitation (meRIP) protocol, as well as a set of in silico experiments. By applying Episo to public human and mouse RNA-BisSeq data, we found that the RNA m5C is not evenly distributed among the transcript isoforms, implying the m5C may subject to be regulated at isoform level. AVAILABILITY: Episo is released under the GNU GPLv3+ license. The resource code Episo is freely accessible from https://github.com/liujunfengtop/Episo (with Tophat/cufflink) and https://github.com/liujunfengtop/Episo/tree/master/Episo_Kallisto (with Kallisto). SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.</p>',
'date' => '2019-12-03',
'pmid' => 'http://www.pubmed.gov/31794005',
'doi' => '10.1093/bioinformatics/btz900/5651015',
'modified' => '2020-02-25 13:26:22',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '3499',
'name' => 'Centromeres License the Mitotic Condensation of Yeast Chromosome Arms.',
'authors' => 'Kruitwagen T, Chymkowitch P, Denoth-Lippuner A, Enserink J, Barral Y',
'description' => '<p>During mitosis, chromatin condensation shapes chromosomes as separate, rigid, and compact sister chromatids to facilitate their segregation. Here, we show that, unlike wild-type yeast chromosomes, non-chromosomal DNA circles and chromosomes lacking a centromere fail to condense during mitosis. The centromere promotes chromosome condensation strictly in cis through recruiting the kinases Aurora B and Bub1, which trigger the autonomous condensation of the entire chromosome. Shugoshin and the deacetylase Hst2 facilitated spreading the condensation signal to the chromosome arms. Targeting Aurora B to DNA circles or centromere-ablated chromosomes or releasing Shugoshin from PP2A-dependent inhibition bypassed the centromere requirement for condensation and enhanced the mitotic stability of DNA circles. Our data indicate that yeast cells license the chromosome-autonomous condensation of their chromatin in a centromere-dependent manner, excluding from this process non-centromeric DNA and thereby inhibiting their propagation.</p>',
'date' => '2018-10-18',
'pmid' => 'http://www.pubmed.org/30318142',
'doi' => '10.1016/j.cell.2018.09.012',
'modified' => '2019-02-27 15:44:25',
'created' => '2019-02-27 12:54:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '3235',
'name' => 'A novel FOXA1/ESR1 interacting pathway: A study of Oncomine™ breast cancer microarrays',
'authors' => 'Chaudhary S. et al.',
'description' => '<p>Forkhead box protein A1 (FOXA1) is essential for the growth and differentiation of breast epithelium, and has a favorable outcome in breast cancer (BC). Elevated <i>FOXA1</i> expression in BC also facilitates hormone responsiveness in estrogen receptor (<i>ESR</i>)-positive BC. However, the interaction between these two pathways is not fully understood. <i>FOXA1</i> and GATA binding protein 3 (<i>GATA3</i>) along with <i>ESR1</i> expression are responsible for maintaining a luminal phenotype, thus suggesting the existence of a strong association between them. The present study utilized the Oncomine™ microarray database to identify <i>FOXA1:ESR1</i> and <i>FOXA1:ESR1:GATA3</i> co-expression co-regulated genes. Oncomine™ analysis revealed 115 and 79 overlapping genes clusters in <i>FOXA1:ESR1</i> and <i>FOXA1:ESR1:GATA3</i> microarrays, respectively. Five ESR1 direct target genes [trefoil factor 1 (<i>TFF1/PS2</i>), <i>B-cell lymphoma</i> 2 (<i>BCL2</i>), seven in absentia homolog 2 (<i>SIAH2</i>), cellular myeloblastosis viral oncogene homolog (<i>CMYB</i>) and progesterone receptor (<i>PGR</i>)] were detected in the co-expression clusters. To further investigate the role of FOXA1 in ESR1-positive cells, MCF7 cells were transfected with a <i>FOXA1</i> expression plasmid, and it was observed that the direct target genes of ESR1 (<i>PS2, BCL2, SIAH2</i> and <i>PGR</i>) were significantly regulated upon transfection. Analysis of one of these target genes, <i>PS2</i>, revealed the presence of two FOXA1 binding sites in the vicinity of the estrogen response element (ERE), which was confirmed by binding assays. Under estrogen stimulation, FOXA1 protein was recruited to the FOXA1 site and could also bind to the ERE site (although in minimal amounts) in the <i>PS2</i> promoter. Co-transfection of <i>FOXA1</i>/<i>ESR1</i> expression plasmids demonstrated a significantly regulation of the target genes identified in the <i>FOXA1</i>/<i>ESR1</i> multi-arrays compared with only <i>FOXA1</i> transfection, which was suggestive of a synergistic effect of <i>ESR1</i> and <i>FOXA1</i> on the target genes. In summary, the present study identified novel <i>FOXA1</i>, <i>ESR1</i> and <i>GATA</i>3 co-expressed genes that may be involved in breast tumorigenesis.</p>',
'date' => '2017-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28789340',
'doi' => '',
'modified' => '2017-08-28 09:30:38',
'created' => '2017-08-28 09:30:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '3236',
'name' => 'TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors',
'authors' => 'Ancey P.B. et al.',
'description' => '<p>Understanding the processes that govern liver progenitor cell differentiation has important implications for the design of strategies targeting chronic liver diseases, whereby regeneration of liver tissue is critical. Although DNA methylation (5mC) and hydroxymethylation (5hmC) are highly dynamic during early embryonic development, less is known about their roles at later stages of differentiation. Using an in vitro model of hepatocyte differentiation, we show here that 5hmC precedes the expression of promoter 1 (P1)-dependent isoforms of HNF4A, a master transcription factor of hepatocyte identity. 5hmC and HNF4A expression from P1 are dependent on ten-eleven translocation (TET) dioxygenases. In turn, the liver pioneer factor FOXA2 is necessary for TET1 binding to the P1 locus. Both FOXA2 and TETs are required for the 5hmC-related switch in HNF4A expression. The epigenetic event identified here may be a key step for the establishment of the hepatocyte program by HNF4A.</p>',
'date' => '2017-07-11',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28648900',
'doi' => '',
'modified' => '2017-08-28 10:24:16',
'created' => '2017-08-28 09:44:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '3113',
'name' => 'Differentiation of Mouse Enteric Nervous System Progenitor Cells is Controlled by Endothelin 3 and Requires Regulation of Ednrb by SOX10 and ZEB2',
'authors' => 'Watanabe Y. et al.',
'description' => '<h2 class="sectionTitle"></h2>
<h3 class="sectionTitle">Background & Aims</h3>
<p>Maintenance and differentiation of progenitor cells in the developing enteric nervous system (ENS) are controlled by molecules such as the signaling protein endothelin 3 (EDN3), its receptor (the endothelin receptor type B, EDNRB), and the transcription factors SRY-box 10 (SOX10) and zinc finger E-box binding homeobox 2 (ZEB2). We used enteric progenitor cell (EPC) cultures and mice to study the roles of these proteins in enteric neurogenesis and their cross regulation.</p>
<h3 class="sectionTitle">Methods</h3>
<p>We performed studies in mice with a <em>Zeb2</em> loss-of-function mutation (<em>Zeb2</em><sup>Δ</sup>) and mice carrying a spontaneous recessive mutation that prevents conversion EDN3 to its active form (<em>Edn3</em><sup><em>ls</em></sup>). EPC cultures issued from embryos that expressed only wild-type <em>Zeb2</em> (<em>Zeb2</em><sup>+/+</sup> EPCs) or were heterozygous for the mutation (<em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs) were exposed to EDN3; we analyzed the effects on cell differentiation using immunocytochemistry. In parallel, <em>Edn3</em><sup><em>ls</em></sup> mice were crossed with <em>Zeb2</em><sup><em>Δ/+</em></sup>mice; intestinal tissues were collected from embryos for immunohistochemical analyses. We investigated regulation of the <em>EDNRB</em> gene in transactivation and chromatin immunoprecipitation assays; results were validated in functional rescue experiments using transgenes expression in EPCs from retroviral vectors.</p>
<h3 class="sectionTitle">Results</h3>
<p><em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs had increased neuronal differentiation compared to <em>Zeb2</em><sup><em>+/+</em></sup> cells. When exposed to EDN3, <em>Zeb2</em><sup>+/+</sup> EPCs continued expression of ZEB2 but did not undergo any neuronal differentiation. Incubation of <em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs with EDN3, on the other hand, resulted in only partial inhibition of neuronal differentiation. This indicated that 2 copies of <em>Zeb2</em> are required for EDN3 to prevent neuronal differentiation. Mice with combined mutations in <em>Zeb2</em> and <em>Edn3</em> (double mutants) had more severe enteric anomalies and increased neuronal differentiation compared to mice with mutations in either gene alone. The transcription factors SOX10 and ZEB2 directly activated the <em>EDNRB</em> promoter. Overexpression of EDNRB in <em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs restored inhibition of neuronal differentiation, similar to incubation of <em>Zeb2</em><sup>+/+</sup> EPCs with EDN3.</p>
<h3 class="sectionTitle">Conclusions</h3>
<p>In studies of cultured EPCs and mice, we found that control of differentiation of mouse enteric nervous system progenitor cells by EDN3 requires regulation of <em>Ednrb</em> expression by SOX10 and ZEB2.</p>',
'date' => '2017-01-05',
'pmid' => 'http://www.gastrojournal.org/article/S0016-5085(17)30002-1/abstract?referrer=http%3A%2F%2Fwww.gastrojournal.org%2Farticle%2FS0016-5085%2817%2930002-1%2Ffulltext',
'doi' => '',
'modified' => '2017-01-06 10:04:00',
'created' => '2017-01-06 10:04:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '3183',
'name' => 'High resolution methylation analysis of the HoxA5 regulatory region in different somatic tissues of laboratory mouse during development',
'authors' => 'Sinha P. et al.',
'description' => '<p>Homeobox genes encode a group of DNA binding regulatory proteins whose key function occurs in the spatial-temporal organization of genome during embryonic development and differentiation. The role of these Hox genes during ontogenesis makes it an important model for research. HoxA5 is a member of Hox gene family playing a central role during axial body patterning and morphogenesis. DNA modification studies have shown that the function of Hox genes is partly governed by the methylation-mediated gene expression regulation. Therefore the study aimed to investigate the role of epigenetic events in regulation of tissue-specific expression pattern of HoxA5 gene during mammalian development. The methodology adopted were sodium bisulfite genomic DNA sequencing, quantitative real-time PCR and chromatin-immunoprecipitation (ChIP). Methylation profiling of HoxA5 gene promoter shows higher methylation in adult as compared to fetus in various somatic tissues of mouse being highest in adult spleen. However q-PCR results show higher expression during fetal stages being highest in fetal intestine followed by brain, liver and spleen. These results clearly indicate a strict correlation between DNA methylation and tissue-specific gene expression. The findings of chromatin-immunoprecipitation (ChIP) have also reinforced that epigenetic event like DNA methylation plays important role in the regulation of tissue specific expression of HoxA5.</p>',
'date' => '2017-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28363633',
'doi' => '',
'modified' => '2017-05-22 09:48:38',
'created' => '2017-05-22 09:48:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '3054',
'name' => 'Overexpression of histone demethylase Fbxl10 leads to enhanced migration in mouse embryonic fibroblasts.',
'authors' => 'Rohde M. et al.',
'description' => '<p>Cell migration is a central process in the development and maintenance of multicellular organisms. Tissue formation during embryonic development, wound healing, immune responses and invasive tumors all require the orchestrated movement of cells to specific locations. Histone demethylase proteins alter transcription by regulating the chromatin state at specific gene loci. FBXL10 is a conserved and ubiquitously expressed member of the JmjC domain-containing histone demethylase family and is implicated in the demethylation of H3K4me3 and H3K36me2 and thereby removing active chromatin marks. However, the physiological role of FBXL10 in vivo remains largely unknown. Therefore, we established an inducible gain of function model to analyze the role of Fbxl10 and compared wild-type with Fbxl10 overexpressing mouse embryonic fibroblasts (MEFs). Our study shows that overexpression of Fbxl10 in MEFs doesn't influence the proliferation capability but leads to an enhanced migration capacity in comparison to wild-type MEFs. Transcriptome and ChIP-seq experiments demonstrated that Fbxl10 binds to genes involved in migration like Areg, Mdk, Lmnb1, Thbs1, Mgp and Cxcl12. Taken together, our results strongly suggest that Fbxl10 plays a critical role in migration by binding to the promoter region of migration-associated genes and thereby might influences cell behaviour to a possibly more aggressive phenotype.</p>',
'date' => '2016-09-17',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27646113',
'doi' => '',
'modified' => '2016-10-24 14:35:45',
'created' => '2016-10-24 14:35:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '2979',
'name' => 'Methylation of the Sox9 and Oct4 promoters and its correlation with gene expression during testicular development in the laboratory mouse',
'authors' => 'Pamnani M et al.',
'description' => '<p>Sox9 and Oct4 are two important regulatory factors involved in mammalian development. Sox9, a member of the group E Sox transcription factor family, has a crucial role in the development of the genitourinary system, while Oct4, commonly known as octamer binding transcription factor 4, belongs to class V of the transcription family. The expression of these two proteins exhibits a dynamic pattern with regard to their expression sites and levels. The aim of this study was to investigate the role of de novo methylation in the regulation of the tissue- and site-specific expression of these proteins. The dynamics of the de novo methylation of 15 CpGs and six CpGs in Sox9 and Oct4 respectively, was studied with sodium bisulfite genomic DNA sequencing in mouse testis at different developmental stages. Consistent methylation of three CpGs was observed in adult ovary in which the expression of Sox9 was feeble, while the level of methylation in somatic tissue was greater in Oct4 compared to germinal tissue. The promoter-chromatin status of Sox9 was also studied with a chromatin immune-precipitation assay.</p>',
'date' => '2016-07-04',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27381637',
'doi' => '10.1590/1678-4685-GMB-2015-0172',
'modified' => '2016-07-11 12:31:08',
'created' => '2016-07-11 12:31:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '2897',
'name' => 'Overexpression of caspase 7 is ERα dependent to affect proliferation and cell growth in breast cancer cells by targeting p21(Cip)',
'authors' => 'Chaudhary S, Madhukrishna B, Adhya AK, Keshari S, Mishra SK',
'description' => '<p>Caspase 7 (CASP7) expression has important function during cell cycle progression and cell growth in certain cancer cells and is also involved in the development and differentiation of dental tissues. However, the function of CASP7 in breast cancer cells is unclear. The aim of this study was to analyze the expression of CASP7 in breast carcinoma patients and determine the role of CASP7 in regulating tumorigenicity in breast cancer cells. In this study, we show that the CASP7 expression is high in breast carcinoma tissues compared with normal counterpart. The ectopic expression of CASP7 is significantly associated with ERα expression status and persistently elevated in different stages of the breast tumor grades. High level of CASP7 expression showed better prognosis in breast cancer patients with systemic endocrine therapy as observed from Kaplan-Meier analysis. S3 and S4, estrogen responsive element (ERE) in the CASP7 promoter, is important for estrogen-ERα-mediated CASP7 overexpression. Increased recruitment of p300, acetylated H3 and pol II in the ERE region of CASP7 promoter is observed after hormone stimulation. Ectopic expression of CASP7 in breast cancer cells results in cell growth and proliferation inhibition via p21(Cip) reduction, whereas small interfering RNA (siRNA) mediated reduction of CASP7 rescued p21(Cip) levels. We also show that pro- and active forms of CASP7 is located in the nucleus apart from cytoplasmic region of breast cancer cells. The proliferation and growth of breast cancer cells is significantly reduced by broad-spectrum peptide inhibitors and siRNA of CASP7. Taken together, our findings show that CASP7 is aberrantly expressed in breast cancer and contributes to cell growth and proliferation by downregulating p21(Cip) protein, suggesting that targeting CASP7-positive breast cancer could be one of the potential therapeutic strategies.</p>',
'date' => '2016-04-18',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27089142',
'doi' => '10.1038/oncsis.2016.12',
'modified' => '2016-04-28 10:15:00',
'created' => '2016-04-28 10:15:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '2834',
'name' => 'Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.',
'authors' => 'Rebollo R, Mager DL',
'description' => '<p>Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method used in our laboratory to detect, quantify, and compare mammalian endogenous retrovirus DNA methylation. More specifically we describe methylated DNA immunoprecipitation (MeDIP) followed by quantitative PCR.</p>',
'date' => '2016-02-20',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26895065',
'doi' => '10.1007/978-1-4939-3372-3_23',
'modified' => '2016-03-02 10:37:15',
'created' => '2016-03-02 10:22:16',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '2842',
'name' => 'Chromatin Immunoprecipitation Assay for the Identification of Arabidopsis Protein-DNA Interactions In Vivo',
'authors' => 'Komar DN, Mouriz A, Jarillo JA, Piñeiro M',
'description' => '<p>Intricate gene regulatory networks orchestrate biological processes and developmental transitions in plants. Selective transcriptional activation and silencing of genes mediate the response of plants to environmental signals and developmental cues. Therefore, insights into the mechanisms that control plant gene expression are essential to gain a deep understanding of how biological processes are regulated in plants. The chromatin immunoprecipitation (ChIP) technique described here is a procedure to identify the DNA-binding sites of proteins in genes or genomic regions of the model species Arabidopsis thaliana. The interactions with DNA of proteins of interest such as transcription factors, chromatin proteins or posttranslationally modified versions of histones can be efficiently analyzed with the ChIP protocol. This method is based on the fixation of protein-DNA interactions in vivo, random fragmentation of chromatin, immunoprecipitation of protein-DNA complexes with specific antibodies, and quantification of the DNA associated with the protein of interest by PCR techniques. The use of this methodology in Arabidopsis has contributed significantly to unveil transcriptional regulatory mechanisms that control a variety of plant biological processes. This approach allowed the identification of the binding sites of the Arabidopsis chromatin protein EBS to regulatory regions of the master gene of flowering FT. The impact of this protein in the accumulation of particular histone marks in the genomic region of FT was also revealed through ChIP analysis.</p>',
'date' => '2016-01-14',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26863263',
'doi' => '10.3791/53422',
'modified' => '2017-01-04 14:16:52',
'created' => '2016-03-09 17:05:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '2801',
'name' => 'The genetic association of RUNX3 with ankylosing spondylitis can be explained by allele-specific effects on IRF4 recruitment that alter gene expression',
'authors' => 'Matteo Vecellio, Amity R Roberts, Carla J Cohen, Adrian Cortes, Julian C Knight, Paul Bowness, B Paul Wordsworth',
'description' => '<p>The authors sought to identify the functional basis for the genetic association of single nucleotide polymorphisms (SNP), upstream of the RUNX3 promoter, with ankylosing spondylitis (AS). They performed conditional analysis of genetic association data and used ENCODE data on chromatin remodelling and transcription factor (TF) binding sites to identify the primary AS-associated regulatory SNP in the RUNX3 region. The functional effects of this SNP were tested in luciferase reporter assays. Its effects on TF binding were investigated by electrophoretic mobility gel shift assays and chromatin immunoprecipitation. RUNX3 mRNA levels were compared in primary CD8+ T cells of AS risk and protective genotypes by real-time PCR. They identified a regulatory region upstream of RUNX3 that is modulated byrs4648889. The risk allele decreases TF binding (including IRF4) and reduces reporter activity and RUNX3 expression. These findings may have important implications for understanding the role of T cells and other immune cells in AS.</p>',
'date' => '2015-10-09',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26452539',
'doi' => '10.1136/annrheumdis-2015-207490',
'modified' => '2016-04-25 09:56:04',
'created' => '2015-12-07 06:37:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '2852',
'name' => 'ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells',
'authors' => 'Al-Furoukh N, Ianni A, Nolte H, Hölper S, Krüger M, Wanrooij S, Braun T',
'description' => '<p>Proteostasis is crucial for life and maintained by cellular chaperones and proteases. One major mitochondrial protease is the ClpXP complex, which is comprised of a catalytic ClpX subunit and a proteolytic ClpP subunit. Based on two separate observations, we hypothesized that ClpX may play a leading role in the cellular function of ClpXP. Therefore, we analyzed the effect of ClpX overexpression on a myoblast proteome by quantitative proteomics. ClpX overexpression results in the upregulation of markers of the mitochondrial proteostasis pathway, known as the "mitochondrial unfolded protein response" (UPRmt). Although this pathway is described in detail in Caenorhabditis elegans, it is not clear whether it is conserved in mammals. Therefore, we compared features of the classical nematode UPRmt with our mammalian ClpX-triggered UPRmt dataset. We show that they share the same retrograde mitochondria-to-nucleus signaling pathway that involves the key UPRmt transcription factor CHOP (also known as Ddit3, CEBPZ or GADD153). In conclusion, our data confirm the existence of a mammalian UPRmt that has great similarity to the C. elegans pathway. Furthermore, our results illustrate that ClpX overexpression is a good and simple model to study the underlying mechanisms of the UPRmt in mammalian cells.</p>',
'date' => '2015-10-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26142927',
'doi' => '10.1016/j.bbamcr.2015.06.016',
'modified' => '2016-03-14 10:20:09',
'created' => '2016-03-14 10:20:09',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '2890',
'name' => 'Endothelial Msx1 transduces hemodynamic changes into an arteriogenic remodeling response.',
'authors' => 'Vandersmissen I, Craps S, Depypere M, Coppiello G, van Gastel N, Maes F, Carmeliet G, Schrooten J, Jones EA, Umans L, Devlieger R, Koole M, Gheysens O, Zwijsen A, Aranguren XL, Luttun A',
'description' => '<p>Collateral remodeling is critical for blood flow restoration in peripheral arterial disease and is triggered by increasing fluid shear stress in preexisting collateral arteries. So far, no arterial-specific mediators of this mechanotransduction response have been identified. We show that muscle segment homeobox 1 (MSX1) acts exclusively in collateral arterial endothelium to transduce the extrinsic shear stimulus into an arteriogenic remodeling response. MSX1 was specifically up-regulated in remodeling collateral arteries. MSX1 induction in collateral endothelial cells (ECs) was shear stress driven and downstream of canonical bone morphogenetic protein-SMAD signaling. Flow recovery and collateral remodeling were significantly blunted in EC-specific Msx1/2 knockout mice. Mechanistically, MSX1 linked the arterial shear stimulus to arteriogenic remodeling by activating the endothelial but not medial layer to a proinflammatory state because EC but not smooth muscle cellMsx1/2 knockout mice had reduced leukocyte recruitment to remodeling collateral arteries. This reduced leukocyte infiltration in EC Msx1/2 knockout mice originated from decreased levels of intercellular adhesion molecule 1 (ICAM1)/vascular cell adhesion molecule 1 (VCAM1), whose expression was also in vitro driven by promoter binding of MSX1.</p>',
'date' => '2015-09-28',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26391659',
'doi' => ' 10.1083/jcb.201502003',
'modified' => '2016-04-12 10:44:22',
'created' => '2016-04-12 10:44:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '2846',
'name' => 'DNA methylation directs functional maturation of pancreatic β cells',
'authors' => 'Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matveyenko A, Bhushan A.',
'description' => '<p>Pancreatic β cells secrete insulin in response to postprandial increases in glucose levels to prevent hyperglycemia and inhibit insulin secretion under fasting conditions to protect against hypoglycemia. β cells lack this functional capability at birth and acquire glucose-stimulated insulin secretion (GSIS) during neonatal life. Here, we have shown that during postnatal life, the de novo DNA methyltransferase DNMT3A initiates a metabolic program by repressing key genes, thereby enabling the coupling of insulin secretion to glucose levels. In a murine model, β cell-specific deletion of Dnmt3a prevented the metabolic switch, resulting in loss of GSIS. DNMT3A bound to the promoters of the genes encoding hexokinase 1 (HK1) and lactate dehydrogenase A (LDHA) - both of which regulate the metabolic switch - and knockdown of these two key DNMT3A targets restored the GSIS response in islets from animals with β cell-specific Dnmt3a deletion. Furthermore, DNA methylation-mediated repression of glucose-secretion decoupling genes to modulate GSIS was conserved in human β cells. Together, our results reveal a role for DNA methylation to direct the acquisition of pancreatic β cell function.</p>',
'date' => '2015-07-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26098213',
'doi' => '10.1172/JCI79956',
'modified' => '2016-03-11 14:16:44',
'created' => '2016-03-11 14:16:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '2049',
'name' => 'Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions.',
'authors' => 'Tapias A, Zhou ZW, Shi Y, Chong Z, Wang P, Groth M, Platzer M, Huttner W, Herceg Z, Yang YG, Wang ZQ',
'description' => 'Fate decisions in neural progenitor cells are orchestrated via multiple pathways, and the role of histone acetylation in these decisions has been ascribed to a general function promoting gene activation. Here, we show that the histone acetyltransferase (HAT) cofactor transformation/transcription domain-associated protein (Trrap) specifically regulates activation of cell-cycle genes, thereby integrating discrete cell-intrinsic programs of cell-cycle progression and epigenetic regulation of gene transcription in order to control neurogenesis. Deletion of Trrap impairs recruitment of HATs and transcriptional machinery specifically to E2F cell-cycle target genes, disrupting their transcription with consequent cell-cycle lengthening specifically within cortical apical neural progenitors (APs). Consistently, Trrap conditional mutants exhibit microcephaly because of premature differentiation of APs into intermediate basal progenitors and neurons, and overexpressing cell-cycle regulators in vivo can rescue these premature differentiation defects. These results demonstrate an essential and highly specific role for Trrap-mediated histone regulation in controlling cell-cycle progression and neurogenesis.',
'date' => '2014-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24792116',
'doi' => '',
'modified' => '2015-07-24 15:39:02',
'created' => '2015-07-24 15:39:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '1493',
'name' => 'Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks.',
'authors' => 'Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gäbel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D',
'description' => 'The chromosome 9p21 (Chr9p21) locus of coronary artery disease has been identified in the first surge of genome-wide association and is the strongest genetic factor of atherosclerosis known today. Chr9p21 encodes the long non-coding RNA (ncRNA) antisense non-coding RNA in the INK4 locus (ANRIL). ANRIL expression is associated with the Chr9p21 genotype and correlated with atherosclerosis severity. Here, we report on the molecular mechanisms through which ANRIL regulates target-genes in trans, leading to increased cell proliferation, increased cell adhesion and decreased apoptosis, which are all essential mechanisms of atherogenesis. Importantly, trans-regulation was dependent on Alu motifs, which marked the promoters of ANRIL target genes and were mirrored in ANRIL RNA transcripts. ANRIL bound Polycomb group proteins that were highly enriched in the proximity of Alu motifs across the genome and were recruited to promoters of target genes upon ANRIL over-expression. The functional relevance of Alu motifs in ANRIL was confirmed by deletion and mutagenesis, reversing trans-regulation and atherogenic cell functions. ANRIL-regulated networks were confirmed in 2280 individuals with and without coronary artery disease and functionally validated in primary cells from patients carrying the Chr9p21 risk allele. Our study provides a molecular mechanism for pro-atherogenic effects of ANRIL at Chr9p21 and suggests a novel role for Alu elements in epigenetic gene regulation by long ncRNAs.',
'date' => '2013-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23861667',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '1285',
'name' => 'Genome-wide analysis of LXRα activation reveals new transcriptional networks in human atherosclerotic foam cells.',
'authors' => 'Feldmann R, Fischer C, Kodelja V, Behrens S, Haas S, Vingron M, Timmermann B, Geikowski A, Sauer S',
'description' => 'Increased physiological levels of oxysterols are major risk factors for developing atherosclerosis and cardiovascular disease. Lipid-loaded macrophages, termed foam cells, are important during the early development of atherosclerotic plaques. To pursue the hypothesis that ligand-based modulation of the nuclear receptor LXRα is crucial for cell homeostasis during atherosclerotic processes, we analysed genome-wide the action of LXRα in foam cells and macrophages. By integrating chromatin immunoprecipitation-sequencing (ChIP-seq) and gene expression profile analyses, we generated a highly stringent set of 186 LXRα target genes. Treatment with the nanomolar-binding ligand T0901317 and subsequent auto-regulatory LXRα activation resulted in sequence-dependent sharpening of the genome-binding patterns of LXRα. LXRα-binding loci that correlated with differential gene expression revealed 32 novel target genes with potential beneficial effects, which in part explained the implications of disease-associated genetic variation data. These observations identified highly integrated LXRα ligand-dependent transcriptional networks, including the APOE/C1/C4/C2-gene cluster, which contribute to the reversal of cholesterol efflux and the dampening of inflammation processes in foam cells to prevent atherogenesis.',
'date' => '2013-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23393188',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '1078',
'name' => 'New partners in regulation of gene expression: the enhancer of trithorax and polycomb corto interacts with methylated ribosomal protein l12 via its chromodomain.',
'authors' => 'Coléno-Costes A, Jang SM, de Vanssay A, Rougeot J, Bouceba T, Randsholt NB, Gibert JM, Le Crom S, Mouchel-Vielh E, Bloyer S, Peronnet F',
'description' => 'Chromodomains are found in many regulators of chromatin structure, and most of them recognize methylated lysines on histones. Here, we investigate the role of the Drosophila melanogaster protein Corto's chromodomain. The Enhancer of Trithorax and Polycomb Corto is involved in both silencing and activation of gene expression. Over-expression of the Corto chromodomain (CortoCD) in transgenic flies shows that it is a chromatin-targeting module, critical for Corto function. Unexpectedly, mass spectrometry analysis reveals that polypeptides pulled down by CortoCD from nuclear extracts correspond to ribosomal proteins. Furthermore, real-time interaction analyses demonstrate that CortoCD binds with high affinity RPL12 tri-methylated on lysine 3. Corto and RPL12 co-localize with active epigenetic marks on polytene chromosomes, suggesting that both are involved in fine-tuning transcription of genes in open chromatin. RNA-seq based transcriptomes of wing imaginal discs over-expressing either CortoCD or RPL12 reveal that both factors deregulate large sets of common genes, which are enriched in heat-response and ribosomal protein genes, suggesting that they could be implicated in dynamic coordination of ribosome biogenesis. Chromatin immunoprecipitation experiments show that Corto and RPL12 bind hsp70 and are similarly recruited on gene body after heat shock. Hence, Corto and RPL12 could be involved together in regulation of gene transcription. We discuss whether pseudo-ribosomal complexes composed of various ribosomal proteins might participate in regulation of gene expression in connection with chromatin regulators.',
'date' => '2012-10-11',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23071455',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '930',
'name' => 'The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts',
'authors' => 'Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J',
'description' => 'Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain and a Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts (MEFs) stably over-expressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the non-coding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2 and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches, showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression and the metabolic control of fibroblasts. ',
'date' => '2012-07-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22825849',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '352',
'name' => 'Mouse IgG SDS US en',
'language' => 'en',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-US-en-GHS_1_0.pdf',
'countries' => 'US',
'modified' => '2020-06-09 15:24:43',
'created' => '2020-06-09 15:24:43',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '350',
'name' => 'Mouse IgG SDS GB en',
'language' => 'en',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-GB-en-GHS_1_0.pdf',
'countries' => 'GB',
'modified' => '2020-06-09 15:23:14',
'created' => '2020-06-09 15:23:14',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '348',
'name' => 'Mouse IgG SDS ES es',
'language' => 'es',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-ES-es-GHS_1_0.pdf',
'countries' => 'ES',
'modified' => '2020-06-09 15:22:03',
'created' => '2020-06-09 15:22:03',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '347',
'name' => 'Mouse IgG SDS DE de',
'language' => 'de',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-DE-de-GHS_1_0.pdf',
'countries' => 'DE',
'modified' => '2020-06-09 15:21:25',
'created' => '2020-06-09 15:21:25',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '351',
'name' => 'Mouse IgG SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-JP-ja-GHS_1_0.pdf',
'countries' => 'JP',
'modified' => '2020-06-09 15:23:45',
'created' => '2020-06-09 15:23:45',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '346',
'name' => 'Mouse IgG SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-BE-nl-GHS_1_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-09 15:20:49',
'created' => '2020-06-09 15:20:49',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '345',
'name' => 'Mouse IgG SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-BE-fr-GHS_1_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-09 15:19:47',
'created' => '2020-06-09 15:19:47',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '349',
'name' => 'Mouse IgG SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-FR-fr-GHS_1_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-09 15:22:38',
'created' => '2020-06-09 15:22:38',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/mouse-igg-15-ug-15-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
)
$pro = array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/bioruptor-pico-sonication-device"><img src="/img/product/shearing_technologies/bioruptor_pico.jpg" alt="Bioruptor pico next gen sequencing " class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">B01060010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Bioruptor® Pico sonication device</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '1862',
'product_id' => '1959',
'related_id' => '1787'
),
'Image' => array(
(int) 0 => array(
'id' => '133',
'name' => 'product/shearing_technologies/bioruptor_pico.jpg',
'alt' => 'Bioruptor pico next gen sequencing ',
'modified' => '2020-11-27 10:00:30',
'created' => '2015-06-10 17:28:55',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1772',
'name' => 'product/shearing_technologies/B010600010.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 15:41:46',
'created' => '2018-02-14 15:41:46',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1773',
'name' => 'product/shearing_technologies/B010600010-2.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 16:18:03',
'created' => '2018-02-14 15:42:00',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1774',
'name' => 'product/shearing_technologies/B01060010-bioruptor-pico-cooler.jpg',
'alt' => 'Pico + cooler',
'modified' => '2018-03-06 11:01:33',
'created' => '2018-03-06 11:01:33',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
'id' => '4167',
'product_id' => '1959',
'application_id' => '43'
)
)
$slugs = array(
(int) 0 => 'chip-qpcr-antibodies'
)
$applications = array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'locale' => 'eng'
)
$description = ''
$name = 'ChIP-qPCR (ab)'
$document = array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '2068',
'product_id' => '1959',
'document_id' => '38'
)
)
$sds = array(
'id' => '349',
'name' => 'Mouse IgG SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-FR-fr-GHS_1_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-09 15:22:38',
'created' => '2020-06-09 15:22:38',
'ProductsSafetySheet' => array(
'id' => '681',
'product_id' => '1959',
'safety_sheet_id' => '349'
)
)
$publication = array(
'id' => '930',
'name' => 'The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts',
'authors' => 'Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J',
'description' => 'Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain and a Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts (MEFs) stably over-expressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the non-coding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2 and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches, showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression and the metabolic control of fibroblasts. ',
'date' => '2012-07-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22825849',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
'id' => '1075',
'product_id' => '1959',
'publication_id' => '930'
)
)
$externalLink = ' <a href="http://www.ncbi.nlm.nih.gov/pubmed/22825849" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: message [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'product' => array(
'Product' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '328',
'name' => 'Mouse IgG',
'description' => 'The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative contro',
'clonality' => '',
'isotype' => '',
'lot' => '',
'concentration' => '1.0 µg/µl',
'reactivity' => 'none',
'type' => 'Polyclonal',
'purity' => 'Protein A purified ',
'classification' => '',
'application_table' => '',
'storage_conditions' => '',
'storage_buffer' => '',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-02-24 12:10:53',
'created' => '0000-00-00 00:00:00',
'select_label' => '328 - Mouse IgG ( - 1.0 µg/µl - none - Protein A purified - Mouse)'
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/mouse-igg-15-ug-15-ul'
)
$language = 'en'
$meta_keywords = ''
$meta_description = 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.'
$meta_title = 'Mouse IgG (C15400001) | Diagenode'
$product = array(
'Product' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '328',
'name' => 'Mouse IgG',
'description' => 'The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative contro',
'clonality' => '',
'isotype' => '',
'lot' => '',
'concentration' => '1.0 µg/µl',
'reactivity' => 'none',
'type' => 'Polyclonal',
'purity' => 'Protein A purified ',
'classification' => '',
'application_table' => '',
'storage_conditions' => '',
'storage_buffer' => '',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-02-24 12:10:53',
'created' => '0000-00-00 00:00:00',
'select_label' => '328 - Mouse IgG ( - 1.0 µg/µl - none - Protein A purified - Mouse)'
),
'Slave' => array(
(int) 0 => array(
'id' => '211',
'name' => 'C15400001',
'product_id' => '1959',
'modified' => '2017-05-17 15:27:44',
'created' => '2017-05-17 15:27:44'
)
),
'Group' => array(
'Group' => array(
'id' => '211',
'name' => 'C15400001',
'product_id' => '1959',
'modified' => '2017-05-17 15:27:44',
'created' => '2017-05-17 15:27:44'
),
'Master' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '7',
'position' => '10',
'parent_id' => '1',
'name' => 'Methylated DNA immunoprecipitation',
'description' => '<div class="row extra-spaced">
<div class="small-12 medium-3 large-3 columns"><center><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank"><img src="https://www.diagenode.com/img/banners/banner-nature-publication-580.png" /></a></center></div>
<div class="small-12 medium-9 large-9 columns">
<h3>Sensitive tumour detection and classification using plasma cell-free DNA methylomes<br /><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank">Read the publication</a></h3>
<h3 class="c-article-title u-h1" data-test="article-title" itemprop="name headline">Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA<br /><a href="https://www.nature.com/articles/s41596-019-0202-2" target="_blank" title="cfMeDIP-seq Nature Method">Read the method</a></h3>
</div>
</div>
<div class="row">
<div class="large-12 columns"><span>The Methylated DNA Immunoprecipitation is based on the affinity purification of methylated and hydroxymethylated DNA using, respectively, an antibody directed against 5-methylcytosine (5-mC) in the case of MeDIP or 5-hydroxymethylcytosine (5-hmC) in the case of hMeDIP.</span><br />
<h2></h2>
<h2>How it works</h2>
<p>In brief, Methyl DNA IP is performed as follows: Genomic DNA from cultured cells or tissues is prepared, sheared, and then denatured. Then, immunoselection and immunoprecipitation can take place using the antibody directed against 5 methylcytosine and antibody binding beads. After isolation and purification is performed, the IP’d methylated DNA is ready for any subsequent analysis as qPCR, amplification, hybridization on microarrays or next generation sequencing.</p>
<h2>Applications</h2>
<div align="center"><a href="https://www.diagenode.com/en/p/magmedip-kit-x48-48-rxns" class="center alert radius button"> qPCR analysis</a></div>
<div align="center"><a href="https://www.diagenode.com/en/p/magmedip-seq-package-V2-x10" class="center alert radius button"> NGS analysis </a></div>
<h2>Advantages</h2>
<ul style="font-size: 19px;" class="nobullet">
<li><i class="fa fa-arrow-circle-right"></i> <strong>Unaffected</strong> DNA</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>High enrichment</strong> yield</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>Robust</strong> & <strong>reproducible</strong> techniques</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>NGS</strong> compatible</li>
</ul>
<h2></h2>
</div>
</div>
<div id="gtx-trans" style="position: absolute; left: 17px; top: 652.938px;">
<div class="gtx-trans-icon"></div>
</div>',
'in_footer' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'slug' => 'methylated-dna-immunoprecipitation',
'meta_keywords' => 'Methylated DNA immunoprecipitation,Epigenetic,DNA Methylation,qPCR,5 methylcytosine (5-mC)',
'meta_description' => 'Methylated DNA immunoprecipitation method is based on the affinity purification of methylated DNA using an antibody directed against 5 methylcytosine (5-mC). ',
'meta_title' => 'Methylated DNA immunoprecipitation(MeDIP) - Dna methylation | Diagenode',
'modified' => '2021-08-19 12:08:03',
'created' => '2014-09-14 05:33:34',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '22',
'position' => '40',
'parent_id' => '4',
'name' => 'Isotype controls',
'description' => '<p><span style="font-weight: 400;">Diagenode offers the negative Ctrl IgG from rabbit, rat and mouse. These extensively validated antibodies can be used as negative controls in ChIP, IF, hMeDIP or other experiments performed with specific antibodies made in rabbit, rat or mouse, respectively.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'isotype-controls',
'cookies_tag_id' => null,
'meta_keywords' => 'Isotype controls,DNA immunoprecipitation,Methylated DNA immunoprecipitation',
'meta_description' => 'Diagenode provides Isotype controls for Methylated DNA immunoprecipitation',
'meta_title' => 'Isotype controls for Methylated DNA immunoprecipitation | Diagenode',
'modified' => '2019-07-04 16:19:36',
'created' => '2014-09-30 14:23:34',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '127',
'position' => '10',
'parent_id' => '4',
'name' => 'ChIP-grade antibodies',
'description' => '<div class="row">
<div class="small-10 columns"><center></center>
<p><br />Chromatin immunoprecipitation (<b>ChIP</b>) is a technique to study the associations of proteins with the specific genomic regions in intact cells. One of the most important steps of this protocol is the immunoprecipitation of targeted protein using the antibody specifically recognizing it. The quality of antibodies used in ChIP is essential for the success of the experiment. Diagenode offers extensively validated ChIP-grade antibodies, confirmed for their specificity, and high level of performance in ChIP. Each batch is validated, and batch-specific data are available on the website.</p>
<p></p>
</div>
<div class="small-2 columns"><img src="https://www.diagenode.com/emailing/images/epi-success-guaranteed-icon.png" alt="Epigenetic success guaranteed" /></div>
</div>
<p><strong>ChIP results</strong> obtained with the antibody directed against H3K4me3 (Cat. No. <a href="../p/h3k4me3-polyclonal-antibody-premium-50-ug-50-ul">C15410003</a>). </p>
<div class="row">
<div class="small-12 medium-6 large-6 columns"><img src="https://www.diagenode.com/img/product/antibodies/C15410003-fig1-ChIP.jpg" alt="" width="400" height="315" /> </div>
<div class="small-12 medium-6 large-6 columns">
<p></p>
<p></p>
<p></p>
</div>
</div>
<p></p>
<p>Our aim at Diagenode is to offer the largest collection of highly specific <strong>ChIP-grade antibodies</strong>. We add new antibodies monthly. Find your ChIP-grade antibody in the list below and check more information about tested applications, extensive validation data, and product information.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'chip-grade-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'ChIP-grade antibodies, polyclonal antibody, monoclonal antibody, Diagenode',
'meta_description' => 'Diagenode Offers Extensively Validated ChIP-Grade Antibodies, Confirmed for their Specificity, and high level of Performance in Chromatin Immunoprecipitation ChIP',
'meta_title' => 'Chromatin immunoprecipitation ChIP-grade antibodies | Diagenode',
'modified' => '2021-07-01 10:22:38',
'created' => '2017-02-14 11:16:04',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '726',
'name' => 'Datasheet mouseIgG C15400001',
'description' => 'Datasheet description',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_mouseIgG_C15400001.pdf',
'slug' => 'datasheet-mouseigg-c15400001',
'meta_keywords' => null,
'meta_description' => null,
'modified' => '2015-07-07 11:47:44',
'created' => '2015-07-07 11:47:44',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '250',
'name' => 'product/antibodies/antibody.png',
'alt' => 'Mouse IgG',
'modified' => '2020-11-27 07:00:09',
'created' => '2015-07-17 10:12:18',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4787',
'name' => 'The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.',
'authors' => 'Garcia-Gomez E. et al.',
'description' => '<p>Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the disease (control group). In addition, eight of the PCOS patients underwent intervention with metformin (1500 mg/day) and a carbohydrate-controlled diet (type and quantity) for three months. Clinical and metabolic parameters were determined, and RT-qPCR and MeDIP-qPCR were used to evaluate gene expression and DNA methylation levels, respectively. Decreased expression levels of , , and genes and increased DNA methylation levels of the promoter were found in the endometrium of PCOS patients compared to controls. After metformin and nutritional intervention, some metabolic and clinical variables improved in PCOS patients. This intervention was associated with increased expression of , , and genes and reduced DNA methylation levels of the promoter in the endometrium of PCOS women. Our preliminary findings suggest that metformin and a carbohydrate-controlled diet improve endometrial function in PCOS patients, partly by modulating DNA methylation of the gene promoter and the expression of genes implicated in endometrial receptivity and insulin signaling.</p>',
'date' => '2023-04-01',
'pmid' => 'https://doi.org/10.3390%2Fijms24076857',
'doi' => '10.3390/ijms24076857',
'modified' => '2023-06-12 08:58:33',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4127',
'name' => 'The histone modification H3K4me3 is altered at the locus in Alzheimer'sdisease brain.',
'authors' => 'Smith, Adam et al.',
'description' => '<p>Several epigenome-wide association studies of DNA methylation have highlighted altered DNA methylation in the gene in Alzheimer's disease (AD) brain samples. However, no study has specifically examined histone modifications in the disease. We use chromatin immunoprecipitation-qPCR to quantify tri-methylation at histone 3 lysine 4 (H3K4me3) and 27 (H3K27me3) in the gene in entorhinal cortex from donors with high (n = 59) or low (n = 29) Alzheimer's disease pathology. We demonstrate decreased levels of H3K4me3, a marker of active gene transcription, with no change in H3K27me3, a marker of inactive genes. H3K4me3 is negatively correlated with DNA methylation in specific regions of the gene. Our study suggests that the gene shows altered epigenetic marks indicative of reduced gene activation in Alzheimer's disease.</p>',
'date' => '2021-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33815817',
'doi' => '10.2144/fsoa-2020-0161',
'modified' => '2021-12-07 10:16:08',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4188',
'name' => 'Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage.',
'authors' => 'Ait-Ammar A. et al.',
'description' => '<p>HIV-1 latency generates reservoirs that prevent viral eradication by the current therapies. To find strategies toward an HIV cure, detailed understandings of the molecular mechanisms underlying establishment and persistence of the reservoirs are needed. The cellular transcription factor KAP1 is known as a potent repressor of gene transcription. Here we report that KAP1 represses HIV-1 gene expression in myeloid cells including microglial cells, the major reservoir of the central nervous system. Mechanistically, KAP1 interacts and colocalizes with the viral transactivator Tat to promote its degradation via the proteasome pathway and repress HIV-1 gene expression. In myeloid models of latent HIV-1 infection, the depletion of KAP1 increased viral gene elongation and reactivated HIV-1 expression. Bound to the latent HIV-1 promoter, KAP1 associates and cooperates with CTIP2, a key epigenetic silencer of HIV-1 expression in microglial cells. In addition, Tat and CTIP2 compete for KAP1 binding suggesting a dynamic modulation of the KAP1 cellular partners upon HIV-1 infection. Altogether, our results suggest that KAP1 contributes to the establishment and the persistence of HIV-1 latency in myeloid cells.</p>',
'date' => '2021-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33514850',
'doi' => '10.1038/s41598-021-82164-w',
'modified' => '2022-01-05 15:08:41',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4082',
'name' => 'p53 directly represses human LINE1 transposons.',
'authors' => 'Tiwari, Bhavana and Jones, Amanda E and Caillet, Candace J and Das, Simantiand Royer, Stephanie K and Abrams, John M',
'description' => '<p>p53 is a potent tumor suppressor and commonly mutated in human cancers. Recently, we demonstrated that p53 genes act to restrict retrotransposons in germline tissues of flies and fish but whether this activity is conserved in somatic human cells is not known. Here we show that p53 constitutively restrains human LINE1s by cooperatively engaging sites in the 5'UTR and stimulating local deposition of repressive histone marks at these transposons. Consistent with this, the elimination of p53 or the removal of corresponding binding sites in LINE1s, prompted these retroelements to become hyperactive. Concurrently, p53 loss instigated chromosomal rearrangements linked to LINE sequences and also provoked inflammatory programs that were dependent on reverse transcriptase produced from LINE1s. Taken together, our observations establish that p53 continuously operates at the LINE1 promoter to restrict autonomous copies of these mobile elements in human cells. Our results further suggest that constitutive restriction of these retroelements may help to explain tumor suppression encoded by p53, since erupting LINE1s produced acute oncogenic threats when p53 was absent.</p>',
'date' => '2020-10-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33060137',
'doi' => '10.1101/gad.343186.120',
'modified' => '2021-03-15 16:59:03',
'created' => '2021-02-18 10:21:53',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '3953',
'name' => 'Attenuated Epigenetic Suppression of Muscle Stem Cell Necroptosis Is Required for Efficient Regeneration of Dystrophic Muscles.',
'authors' => 'Sreenivasan K, Ianni A, Künne C, Strilic B, Günther S, Perdiguero E, Krüger M, Spuler S, Offermanns S, Gómez-Del Arco P, Redondo JM, Munoz-Canoves P, Kim J, Braun T',
'description' => '<p>Somatic stem cells expand massively during tissue regeneration, which might require control of cell fitness, allowing elimination of non-competitive, potentially harmful cells. How or if such cells are removed to restore organ function is not fully understood. Here, we show that a substantial fraction of muscle stem cells (MuSCs) undergo necroptosis because of epigenetic rewiring during chronic skeletal muscle regeneration, which is required for efficient regeneration of dystrophic muscles. Inhibition of necroptosis strongly enhances suppression of MuSC expansion in a non-cell-autonomous manner. Prevention of necroptosis in MuSCs of healthy muscles is mediated by the chromatin remodeler CHD4, which directly represses the necroptotic effector Ripk3, while CHD4-dependent Ripk3 repression is dramatically attenuated in dystrophic muscles. Loss of Ripk3 repression by inactivation of Chd4 causes massive necroptosis of MuSCs, abolishing regeneration. Our study demonstrates how programmed cell death in MuSCs is tightly controlled to achieve optimal tissue regeneration.</p>',
'date' => '2020-05-19',
'pmid' => 'http://www.pubmed.gov/32433961',
'doi' => '10.1016/j.celrep.2020.107652',
'modified' => '2020-08-17 09:51:58',
'created' => '2020-08-10 12:12:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '3834',
'name' => 'Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.',
'authors' => 'Liu J, An Z, Luo J, Li J, Li F, Zhang Z',
'description' => '<p>MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of transcriptome-wide profile and potential function of m5C in splicing remains to be elucidated due to lack of isoform level m5C quantification tool. RESULTS: We developed a computational package to quantify Epitranscriptomal RNA m5C at the transcript isoform level (named Episo). Episo consists of three tools, mapper, quant and Bisulfitefq, for mapping, quantifying, and simulating RNA-BisSeq data, respectively. The high accuracy of Episo was validated using an improved m5C-specific methylated RNA immunoprecipitation (meRIP) protocol, as well as a set of in silico experiments. By applying Episo to public human and mouse RNA-BisSeq data, we found that the RNA m5C is not evenly distributed among the transcript isoforms, implying the m5C may subject to be regulated at isoform level. AVAILABILITY: Episo is released under the GNU GPLv3+ license. The resource code Episo is freely accessible from https://github.com/liujunfengtop/Episo (with Tophat/cufflink) and https://github.com/liujunfengtop/Episo/tree/master/Episo_Kallisto (with Kallisto). SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.</p>',
'date' => '2019-12-03',
'pmid' => 'http://www.pubmed.gov/31794005',
'doi' => '10.1093/bioinformatics/btz900/5651015',
'modified' => '2020-02-25 13:26:22',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '3499',
'name' => 'Centromeres License the Mitotic Condensation of Yeast Chromosome Arms.',
'authors' => 'Kruitwagen T, Chymkowitch P, Denoth-Lippuner A, Enserink J, Barral Y',
'description' => '<p>During mitosis, chromatin condensation shapes chromosomes as separate, rigid, and compact sister chromatids to facilitate their segregation. Here, we show that, unlike wild-type yeast chromosomes, non-chromosomal DNA circles and chromosomes lacking a centromere fail to condense during mitosis. The centromere promotes chromosome condensation strictly in cis through recruiting the kinases Aurora B and Bub1, which trigger the autonomous condensation of the entire chromosome. Shugoshin and the deacetylase Hst2 facilitated spreading the condensation signal to the chromosome arms. Targeting Aurora B to DNA circles or centromere-ablated chromosomes or releasing Shugoshin from PP2A-dependent inhibition bypassed the centromere requirement for condensation and enhanced the mitotic stability of DNA circles. Our data indicate that yeast cells license the chromosome-autonomous condensation of their chromatin in a centromere-dependent manner, excluding from this process non-centromeric DNA and thereby inhibiting their propagation.</p>',
'date' => '2018-10-18',
'pmid' => 'http://www.pubmed.org/30318142',
'doi' => '10.1016/j.cell.2018.09.012',
'modified' => '2019-02-27 15:44:25',
'created' => '2019-02-27 12:54:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '3235',
'name' => 'A novel FOXA1/ESR1 interacting pathway: A study of Oncomine™ breast cancer microarrays',
'authors' => 'Chaudhary S. et al.',
'description' => '<p>Forkhead box protein A1 (FOXA1) is essential for the growth and differentiation of breast epithelium, and has a favorable outcome in breast cancer (BC). Elevated <i>FOXA1</i> expression in BC also facilitates hormone responsiveness in estrogen receptor (<i>ESR</i>)-positive BC. However, the interaction between these two pathways is not fully understood. <i>FOXA1</i> and GATA binding protein 3 (<i>GATA3</i>) along with <i>ESR1</i> expression are responsible for maintaining a luminal phenotype, thus suggesting the existence of a strong association between them. The present study utilized the Oncomine™ microarray database to identify <i>FOXA1:ESR1</i> and <i>FOXA1:ESR1:GATA3</i> co-expression co-regulated genes. Oncomine™ analysis revealed 115 and 79 overlapping genes clusters in <i>FOXA1:ESR1</i> and <i>FOXA1:ESR1:GATA3</i> microarrays, respectively. Five ESR1 direct target genes [trefoil factor 1 (<i>TFF1/PS2</i>), <i>B-cell lymphoma</i> 2 (<i>BCL2</i>), seven in absentia homolog 2 (<i>SIAH2</i>), cellular myeloblastosis viral oncogene homolog (<i>CMYB</i>) and progesterone receptor (<i>PGR</i>)] were detected in the co-expression clusters. To further investigate the role of FOXA1 in ESR1-positive cells, MCF7 cells were transfected with a <i>FOXA1</i> expression plasmid, and it was observed that the direct target genes of ESR1 (<i>PS2, BCL2, SIAH2</i> and <i>PGR</i>) were significantly regulated upon transfection. Analysis of one of these target genes, <i>PS2</i>, revealed the presence of two FOXA1 binding sites in the vicinity of the estrogen response element (ERE), which was confirmed by binding assays. Under estrogen stimulation, FOXA1 protein was recruited to the FOXA1 site and could also bind to the ERE site (although in minimal amounts) in the <i>PS2</i> promoter. Co-transfection of <i>FOXA1</i>/<i>ESR1</i> expression plasmids demonstrated a significantly regulation of the target genes identified in the <i>FOXA1</i>/<i>ESR1</i> multi-arrays compared with only <i>FOXA1</i> transfection, which was suggestive of a synergistic effect of <i>ESR1</i> and <i>FOXA1</i> on the target genes. In summary, the present study identified novel <i>FOXA1</i>, <i>ESR1</i> and <i>GATA</i>3 co-expressed genes that may be involved in breast tumorigenesis.</p>',
'date' => '2017-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28789340',
'doi' => '',
'modified' => '2017-08-28 09:30:38',
'created' => '2017-08-28 09:30:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '3236',
'name' => 'TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors',
'authors' => 'Ancey P.B. et al.',
'description' => '<p>Understanding the processes that govern liver progenitor cell differentiation has important implications for the design of strategies targeting chronic liver diseases, whereby regeneration of liver tissue is critical. Although DNA methylation (5mC) and hydroxymethylation (5hmC) are highly dynamic during early embryonic development, less is known about their roles at later stages of differentiation. Using an in vitro model of hepatocyte differentiation, we show here that 5hmC precedes the expression of promoter 1 (P1)-dependent isoforms of HNF4A, a master transcription factor of hepatocyte identity. 5hmC and HNF4A expression from P1 are dependent on ten-eleven translocation (TET) dioxygenases. In turn, the liver pioneer factor FOXA2 is necessary for TET1 binding to the P1 locus. Both FOXA2 and TETs are required for the 5hmC-related switch in HNF4A expression. The epigenetic event identified here may be a key step for the establishment of the hepatocyte program by HNF4A.</p>',
'date' => '2017-07-11',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28648900',
'doi' => '',
'modified' => '2017-08-28 10:24:16',
'created' => '2017-08-28 09:44:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '3113',
'name' => 'Differentiation of Mouse Enteric Nervous System Progenitor Cells is Controlled by Endothelin 3 and Requires Regulation of Ednrb by SOX10 and ZEB2',
'authors' => 'Watanabe Y. et al.',
'description' => '<h2 class="sectionTitle"></h2>
<h3 class="sectionTitle">Background & Aims</h3>
<p>Maintenance and differentiation of progenitor cells in the developing enteric nervous system (ENS) are controlled by molecules such as the signaling protein endothelin 3 (EDN3), its receptor (the endothelin receptor type B, EDNRB), and the transcription factors SRY-box 10 (SOX10) and zinc finger E-box binding homeobox 2 (ZEB2). We used enteric progenitor cell (EPC) cultures and mice to study the roles of these proteins in enteric neurogenesis and their cross regulation.</p>
<h3 class="sectionTitle">Methods</h3>
<p>We performed studies in mice with a <em>Zeb2</em> loss-of-function mutation (<em>Zeb2</em><sup>Δ</sup>) and mice carrying a spontaneous recessive mutation that prevents conversion EDN3 to its active form (<em>Edn3</em><sup><em>ls</em></sup>). EPC cultures issued from embryos that expressed only wild-type <em>Zeb2</em> (<em>Zeb2</em><sup>+/+</sup> EPCs) or were heterozygous for the mutation (<em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs) were exposed to EDN3; we analyzed the effects on cell differentiation using immunocytochemistry. In parallel, <em>Edn3</em><sup><em>ls</em></sup> mice were crossed with <em>Zeb2</em><sup><em>Δ/+</em></sup>mice; intestinal tissues were collected from embryos for immunohistochemical analyses. We investigated regulation of the <em>EDNRB</em> gene in transactivation and chromatin immunoprecipitation assays; results were validated in functional rescue experiments using transgenes expression in EPCs from retroviral vectors.</p>
<h3 class="sectionTitle">Results</h3>
<p><em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs had increased neuronal differentiation compared to <em>Zeb2</em><sup><em>+/+</em></sup> cells. When exposed to EDN3, <em>Zeb2</em><sup>+/+</sup> EPCs continued expression of ZEB2 but did not undergo any neuronal differentiation. Incubation of <em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs with EDN3, on the other hand, resulted in only partial inhibition of neuronal differentiation. This indicated that 2 copies of <em>Zeb2</em> are required for EDN3 to prevent neuronal differentiation. Mice with combined mutations in <em>Zeb2</em> and <em>Edn3</em> (double mutants) had more severe enteric anomalies and increased neuronal differentiation compared to mice with mutations in either gene alone. The transcription factors SOX10 and ZEB2 directly activated the <em>EDNRB</em> promoter. Overexpression of EDNRB in <em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs restored inhibition of neuronal differentiation, similar to incubation of <em>Zeb2</em><sup>+/+</sup> EPCs with EDN3.</p>
<h3 class="sectionTitle">Conclusions</h3>
<p>In studies of cultured EPCs and mice, we found that control of differentiation of mouse enteric nervous system progenitor cells by EDN3 requires regulation of <em>Ednrb</em> expression by SOX10 and ZEB2.</p>',
'date' => '2017-01-05',
'pmid' => 'http://www.gastrojournal.org/article/S0016-5085(17)30002-1/abstract?referrer=http%3A%2F%2Fwww.gastrojournal.org%2Farticle%2FS0016-5085%2817%2930002-1%2Ffulltext',
'doi' => '',
'modified' => '2017-01-06 10:04:00',
'created' => '2017-01-06 10:04:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '3183',
'name' => 'High resolution methylation analysis of the HoxA5 regulatory region in different somatic tissues of laboratory mouse during development',
'authors' => 'Sinha P. et al.',
'description' => '<p>Homeobox genes encode a group of DNA binding regulatory proteins whose key function occurs in the spatial-temporal organization of genome during embryonic development and differentiation. The role of these Hox genes during ontogenesis makes it an important model for research. HoxA5 is a member of Hox gene family playing a central role during axial body patterning and morphogenesis. DNA modification studies have shown that the function of Hox genes is partly governed by the methylation-mediated gene expression regulation. Therefore the study aimed to investigate the role of epigenetic events in regulation of tissue-specific expression pattern of HoxA5 gene during mammalian development. The methodology adopted were sodium bisulfite genomic DNA sequencing, quantitative real-time PCR and chromatin-immunoprecipitation (ChIP). Methylation profiling of HoxA5 gene promoter shows higher methylation in adult as compared to fetus in various somatic tissues of mouse being highest in adult spleen. However q-PCR results show higher expression during fetal stages being highest in fetal intestine followed by brain, liver and spleen. These results clearly indicate a strict correlation between DNA methylation and tissue-specific gene expression. The findings of chromatin-immunoprecipitation (ChIP) have also reinforced that epigenetic event like DNA methylation plays important role in the regulation of tissue specific expression of HoxA5.</p>',
'date' => '2017-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28363633',
'doi' => '',
'modified' => '2017-05-22 09:48:38',
'created' => '2017-05-22 09:48:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '3054',
'name' => 'Overexpression of histone demethylase Fbxl10 leads to enhanced migration in mouse embryonic fibroblasts.',
'authors' => 'Rohde M. et al.',
'description' => '<p>Cell migration is a central process in the development and maintenance of multicellular organisms. Tissue formation during embryonic development, wound healing, immune responses and invasive tumors all require the orchestrated movement of cells to specific locations. Histone demethylase proteins alter transcription by regulating the chromatin state at specific gene loci. FBXL10 is a conserved and ubiquitously expressed member of the JmjC domain-containing histone demethylase family and is implicated in the demethylation of H3K4me3 and H3K36me2 and thereby removing active chromatin marks. However, the physiological role of FBXL10 in vivo remains largely unknown. Therefore, we established an inducible gain of function model to analyze the role of Fbxl10 and compared wild-type with Fbxl10 overexpressing mouse embryonic fibroblasts (MEFs). Our study shows that overexpression of Fbxl10 in MEFs doesn't influence the proliferation capability but leads to an enhanced migration capacity in comparison to wild-type MEFs. Transcriptome and ChIP-seq experiments demonstrated that Fbxl10 binds to genes involved in migration like Areg, Mdk, Lmnb1, Thbs1, Mgp and Cxcl12. Taken together, our results strongly suggest that Fbxl10 plays a critical role in migration by binding to the promoter region of migration-associated genes and thereby might influences cell behaviour to a possibly more aggressive phenotype.</p>',
'date' => '2016-09-17',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27646113',
'doi' => '',
'modified' => '2016-10-24 14:35:45',
'created' => '2016-10-24 14:35:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '2979',
'name' => 'Methylation of the Sox9 and Oct4 promoters and its correlation with gene expression during testicular development in the laboratory mouse',
'authors' => 'Pamnani M et al.',
'description' => '<p>Sox9 and Oct4 are two important regulatory factors involved in mammalian development. Sox9, a member of the group E Sox transcription factor family, has a crucial role in the development of the genitourinary system, while Oct4, commonly known as octamer binding transcription factor 4, belongs to class V of the transcription family. The expression of these two proteins exhibits a dynamic pattern with regard to their expression sites and levels. The aim of this study was to investigate the role of de novo methylation in the regulation of the tissue- and site-specific expression of these proteins. The dynamics of the de novo methylation of 15 CpGs and six CpGs in Sox9 and Oct4 respectively, was studied with sodium bisulfite genomic DNA sequencing in mouse testis at different developmental stages. Consistent methylation of three CpGs was observed in adult ovary in which the expression of Sox9 was feeble, while the level of methylation in somatic tissue was greater in Oct4 compared to germinal tissue. The promoter-chromatin status of Sox9 was also studied with a chromatin immune-precipitation assay.</p>',
'date' => '2016-07-04',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27381637',
'doi' => '10.1590/1678-4685-GMB-2015-0172',
'modified' => '2016-07-11 12:31:08',
'created' => '2016-07-11 12:31:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '2897',
'name' => 'Overexpression of caspase 7 is ERα dependent to affect proliferation and cell growth in breast cancer cells by targeting p21(Cip)',
'authors' => 'Chaudhary S, Madhukrishna B, Adhya AK, Keshari S, Mishra SK',
'description' => '<p>Caspase 7 (CASP7) expression has important function during cell cycle progression and cell growth in certain cancer cells and is also involved in the development and differentiation of dental tissues. However, the function of CASP7 in breast cancer cells is unclear. The aim of this study was to analyze the expression of CASP7 in breast carcinoma patients and determine the role of CASP7 in regulating tumorigenicity in breast cancer cells. In this study, we show that the CASP7 expression is high in breast carcinoma tissues compared with normal counterpart. The ectopic expression of CASP7 is significantly associated with ERα expression status and persistently elevated in different stages of the breast tumor grades. High level of CASP7 expression showed better prognosis in breast cancer patients with systemic endocrine therapy as observed from Kaplan-Meier analysis. S3 and S4, estrogen responsive element (ERE) in the CASP7 promoter, is important for estrogen-ERα-mediated CASP7 overexpression. Increased recruitment of p300, acetylated H3 and pol II in the ERE region of CASP7 promoter is observed after hormone stimulation. Ectopic expression of CASP7 in breast cancer cells results in cell growth and proliferation inhibition via p21(Cip) reduction, whereas small interfering RNA (siRNA) mediated reduction of CASP7 rescued p21(Cip) levels. We also show that pro- and active forms of CASP7 is located in the nucleus apart from cytoplasmic region of breast cancer cells. The proliferation and growth of breast cancer cells is significantly reduced by broad-spectrum peptide inhibitors and siRNA of CASP7. Taken together, our findings show that CASP7 is aberrantly expressed in breast cancer and contributes to cell growth and proliferation by downregulating p21(Cip) protein, suggesting that targeting CASP7-positive breast cancer could be one of the potential therapeutic strategies.</p>',
'date' => '2016-04-18',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27089142',
'doi' => '10.1038/oncsis.2016.12',
'modified' => '2016-04-28 10:15:00',
'created' => '2016-04-28 10:15:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '2834',
'name' => 'Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.',
'authors' => 'Rebollo R, Mager DL',
'description' => '<p>Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method used in our laboratory to detect, quantify, and compare mammalian endogenous retrovirus DNA methylation. More specifically we describe methylated DNA immunoprecipitation (MeDIP) followed by quantitative PCR.</p>',
'date' => '2016-02-20',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26895065',
'doi' => '10.1007/978-1-4939-3372-3_23',
'modified' => '2016-03-02 10:37:15',
'created' => '2016-03-02 10:22:16',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '2842',
'name' => 'Chromatin Immunoprecipitation Assay for the Identification of Arabidopsis Protein-DNA Interactions In Vivo',
'authors' => 'Komar DN, Mouriz A, Jarillo JA, Piñeiro M',
'description' => '<p>Intricate gene regulatory networks orchestrate biological processes and developmental transitions in plants. Selective transcriptional activation and silencing of genes mediate the response of plants to environmental signals and developmental cues. Therefore, insights into the mechanisms that control plant gene expression are essential to gain a deep understanding of how biological processes are regulated in plants. The chromatin immunoprecipitation (ChIP) technique described here is a procedure to identify the DNA-binding sites of proteins in genes or genomic regions of the model species Arabidopsis thaliana. The interactions with DNA of proteins of interest such as transcription factors, chromatin proteins or posttranslationally modified versions of histones can be efficiently analyzed with the ChIP protocol. This method is based on the fixation of protein-DNA interactions in vivo, random fragmentation of chromatin, immunoprecipitation of protein-DNA complexes with specific antibodies, and quantification of the DNA associated with the protein of interest by PCR techniques. The use of this methodology in Arabidopsis has contributed significantly to unveil transcriptional regulatory mechanisms that control a variety of plant biological processes. This approach allowed the identification of the binding sites of the Arabidopsis chromatin protein EBS to regulatory regions of the master gene of flowering FT. The impact of this protein in the accumulation of particular histone marks in the genomic region of FT was also revealed through ChIP analysis.</p>',
'date' => '2016-01-14',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26863263',
'doi' => '10.3791/53422',
'modified' => '2017-01-04 14:16:52',
'created' => '2016-03-09 17:05:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '2801',
'name' => 'The genetic association of RUNX3 with ankylosing spondylitis can be explained by allele-specific effects on IRF4 recruitment that alter gene expression',
'authors' => 'Matteo Vecellio, Amity R Roberts, Carla J Cohen, Adrian Cortes, Julian C Knight, Paul Bowness, B Paul Wordsworth',
'description' => '<p>The authors sought to identify the functional basis for the genetic association of single nucleotide polymorphisms (SNP), upstream of the RUNX3 promoter, with ankylosing spondylitis (AS). They performed conditional analysis of genetic association data and used ENCODE data on chromatin remodelling and transcription factor (TF) binding sites to identify the primary AS-associated regulatory SNP in the RUNX3 region. The functional effects of this SNP were tested in luciferase reporter assays. Its effects on TF binding were investigated by electrophoretic mobility gel shift assays and chromatin immunoprecipitation. RUNX3 mRNA levels were compared in primary CD8+ T cells of AS risk and protective genotypes by real-time PCR. They identified a regulatory region upstream of RUNX3 that is modulated byrs4648889. The risk allele decreases TF binding (including IRF4) and reduces reporter activity and RUNX3 expression. These findings may have important implications for understanding the role of T cells and other immune cells in AS.</p>',
'date' => '2015-10-09',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26452539',
'doi' => '10.1136/annrheumdis-2015-207490',
'modified' => '2016-04-25 09:56:04',
'created' => '2015-12-07 06:37:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '2852',
'name' => 'ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells',
'authors' => 'Al-Furoukh N, Ianni A, Nolte H, Hölper S, Krüger M, Wanrooij S, Braun T',
'description' => '<p>Proteostasis is crucial for life and maintained by cellular chaperones and proteases. One major mitochondrial protease is the ClpXP complex, which is comprised of a catalytic ClpX subunit and a proteolytic ClpP subunit. Based on two separate observations, we hypothesized that ClpX may play a leading role in the cellular function of ClpXP. Therefore, we analyzed the effect of ClpX overexpression on a myoblast proteome by quantitative proteomics. ClpX overexpression results in the upregulation of markers of the mitochondrial proteostasis pathway, known as the "mitochondrial unfolded protein response" (UPRmt). Although this pathway is described in detail in Caenorhabditis elegans, it is not clear whether it is conserved in mammals. Therefore, we compared features of the classical nematode UPRmt with our mammalian ClpX-triggered UPRmt dataset. We show that they share the same retrograde mitochondria-to-nucleus signaling pathway that involves the key UPRmt transcription factor CHOP (also known as Ddit3, CEBPZ or GADD153). In conclusion, our data confirm the existence of a mammalian UPRmt that has great similarity to the C. elegans pathway. Furthermore, our results illustrate that ClpX overexpression is a good and simple model to study the underlying mechanisms of the UPRmt in mammalian cells.</p>',
'date' => '2015-10-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26142927',
'doi' => '10.1016/j.bbamcr.2015.06.016',
'modified' => '2016-03-14 10:20:09',
'created' => '2016-03-14 10:20:09',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '2890',
'name' => 'Endothelial Msx1 transduces hemodynamic changes into an arteriogenic remodeling response.',
'authors' => 'Vandersmissen I, Craps S, Depypere M, Coppiello G, van Gastel N, Maes F, Carmeliet G, Schrooten J, Jones EA, Umans L, Devlieger R, Koole M, Gheysens O, Zwijsen A, Aranguren XL, Luttun A',
'description' => '<p>Collateral remodeling is critical for blood flow restoration in peripheral arterial disease and is triggered by increasing fluid shear stress in preexisting collateral arteries. So far, no arterial-specific mediators of this mechanotransduction response have been identified. We show that muscle segment homeobox 1 (MSX1) acts exclusively in collateral arterial endothelium to transduce the extrinsic shear stimulus into an arteriogenic remodeling response. MSX1 was specifically up-regulated in remodeling collateral arteries. MSX1 induction in collateral endothelial cells (ECs) was shear stress driven and downstream of canonical bone morphogenetic protein-SMAD signaling. Flow recovery and collateral remodeling were significantly blunted in EC-specific Msx1/2 knockout mice. Mechanistically, MSX1 linked the arterial shear stimulus to arteriogenic remodeling by activating the endothelial but not medial layer to a proinflammatory state because EC but not smooth muscle cellMsx1/2 knockout mice had reduced leukocyte recruitment to remodeling collateral arteries. This reduced leukocyte infiltration in EC Msx1/2 knockout mice originated from decreased levels of intercellular adhesion molecule 1 (ICAM1)/vascular cell adhesion molecule 1 (VCAM1), whose expression was also in vitro driven by promoter binding of MSX1.</p>',
'date' => '2015-09-28',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26391659',
'doi' => ' 10.1083/jcb.201502003',
'modified' => '2016-04-12 10:44:22',
'created' => '2016-04-12 10:44:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '2846',
'name' => 'DNA methylation directs functional maturation of pancreatic β cells',
'authors' => 'Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matveyenko A, Bhushan A.',
'description' => '<p>Pancreatic β cells secrete insulin in response to postprandial increases in glucose levels to prevent hyperglycemia and inhibit insulin secretion under fasting conditions to protect against hypoglycemia. β cells lack this functional capability at birth and acquire glucose-stimulated insulin secretion (GSIS) during neonatal life. Here, we have shown that during postnatal life, the de novo DNA methyltransferase DNMT3A initiates a metabolic program by repressing key genes, thereby enabling the coupling of insulin secretion to glucose levels. In a murine model, β cell-specific deletion of Dnmt3a prevented the metabolic switch, resulting in loss of GSIS. DNMT3A bound to the promoters of the genes encoding hexokinase 1 (HK1) and lactate dehydrogenase A (LDHA) - both of which regulate the metabolic switch - and knockdown of these two key DNMT3A targets restored the GSIS response in islets from animals with β cell-specific Dnmt3a deletion. Furthermore, DNA methylation-mediated repression of glucose-secretion decoupling genes to modulate GSIS was conserved in human β cells. Together, our results reveal a role for DNA methylation to direct the acquisition of pancreatic β cell function.</p>',
'date' => '2015-07-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26098213',
'doi' => '10.1172/JCI79956',
'modified' => '2016-03-11 14:16:44',
'created' => '2016-03-11 14:16:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '2049',
'name' => 'Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions.',
'authors' => 'Tapias A, Zhou ZW, Shi Y, Chong Z, Wang P, Groth M, Platzer M, Huttner W, Herceg Z, Yang YG, Wang ZQ',
'description' => 'Fate decisions in neural progenitor cells are orchestrated via multiple pathways, and the role of histone acetylation in these decisions has been ascribed to a general function promoting gene activation. Here, we show that the histone acetyltransferase (HAT) cofactor transformation/transcription domain-associated protein (Trrap) specifically regulates activation of cell-cycle genes, thereby integrating discrete cell-intrinsic programs of cell-cycle progression and epigenetic regulation of gene transcription in order to control neurogenesis. Deletion of Trrap impairs recruitment of HATs and transcriptional machinery specifically to E2F cell-cycle target genes, disrupting their transcription with consequent cell-cycle lengthening specifically within cortical apical neural progenitors (APs). Consistently, Trrap conditional mutants exhibit microcephaly because of premature differentiation of APs into intermediate basal progenitors and neurons, and overexpressing cell-cycle regulators in vivo can rescue these premature differentiation defects. These results demonstrate an essential and highly specific role for Trrap-mediated histone regulation in controlling cell-cycle progression and neurogenesis.',
'date' => '2014-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24792116',
'doi' => '',
'modified' => '2015-07-24 15:39:02',
'created' => '2015-07-24 15:39:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '1493',
'name' => 'Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks.',
'authors' => 'Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gäbel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D',
'description' => 'The chromosome 9p21 (Chr9p21) locus of coronary artery disease has been identified in the first surge of genome-wide association and is the strongest genetic factor of atherosclerosis known today. Chr9p21 encodes the long non-coding RNA (ncRNA) antisense non-coding RNA in the INK4 locus (ANRIL). ANRIL expression is associated with the Chr9p21 genotype and correlated with atherosclerosis severity. Here, we report on the molecular mechanisms through which ANRIL regulates target-genes in trans, leading to increased cell proliferation, increased cell adhesion and decreased apoptosis, which are all essential mechanisms of atherogenesis. Importantly, trans-regulation was dependent on Alu motifs, which marked the promoters of ANRIL target genes and were mirrored in ANRIL RNA transcripts. ANRIL bound Polycomb group proteins that were highly enriched in the proximity of Alu motifs across the genome and were recruited to promoters of target genes upon ANRIL over-expression. The functional relevance of Alu motifs in ANRIL was confirmed by deletion and mutagenesis, reversing trans-regulation and atherogenic cell functions. ANRIL-regulated networks were confirmed in 2280 individuals with and without coronary artery disease and functionally validated in primary cells from patients carrying the Chr9p21 risk allele. Our study provides a molecular mechanism for pro-atherogenic effects of ANRIL at Chr9p21 and suggests a novel role for Alu elements in epigenetic gene regulation by long ncRNAs.',
'date' => '2013-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23861667',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '1285',
'name' => 'Genome-wide analysis of LXRα activation reveals new transcriptional networks in human atherosclerotic foam cells.',
'authors' => 'Feldmann R, Fischer C, Kodelja V, Behrens S, Haas S, Vingron M, Timmermann B, Geikowski A, Sauer S',
'description' => 'Increased physiological levels of oxysterols are major risk factors for developing atherosclerosis and cardiovascular disease. Lipid-loaded macrophages, termed foam cells, are important during the early development of atherosclerotic plaques. To pursue the hypothesis that ligand-based modulation of the nuclear receptor LXRα is crucial for cell homeostasis during atherosclerotic processes, we analysed genome-wide the action of LXRα in foam cells and macrophages. By integrating chromatin immunoprecipitation-sequencing (ChIP-seq) and gene expression profile analyses, we generated a highly stringent set of 186 LXRα target genes. Treatment with the nanomolar-binding ligand T0901317 and subsequent auto-regulatory LXRα activation resulted in sequence-dependent sharpening of the genome-binding patterns of LXRα. LXRα-binding loci that correlated with differential gene expression revealed 32 novel target genes with potential beneficial effects, which in part explained the implications of disease-associated genetic variation data. These observations identified highly integrated LXRα ligand-dependent transcriptional networks, including the APOE/C1/C4/C2-gene cluster, which contribute to the reversal of cholesterol efflux and the dampening of inflammation processes in foam cells to prevent atherogenesis.',
'date' => '2013-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23393188',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '1078',
'name' => 'New partners in regulation of gene expression: the enhancer of trithorax and polycomb corto interacts with methylated ribosomal protein l12 via its chromodomain.',
'authors' => 'Coléno-Costes A, Jang SM, de Vanssay A, Rougeot J, Bouceba T, Randsholt NB, Gibert JM, Le Crom S, Mouchel-Vielh E, Bloyer S, Peronnet F',
'description' => 'Chromodomains are found in many regulators of chromatin structure, and most of them recognize methylated lysines on histones. Here, we investigate the role of the Drosophila melanogaster protein Corto's chromodomain. The Enhancer of Trithorax and Polycomb Corto is involved in both silencing and activation of gene expression. Over-expression of the Corto chromodomain (CortoCD) in transgenic flies shows that it is a chromatin-targeting module, critical for Corto function. Unexpectedly, mass spectrometry analysis reveals that polypeptides pulled down by CortoCD from nuclear extracts correspond to ribosomal proteins. Furthermore, real-time interaction analyses demonstrate that CortoCD binds with high affinity RPL12 tri-methylated on lysine 3. Corto and RPL12 co-localize with active epigenetic marks on polytene chromosomes, suggesting that both are involved in fine-tuning transcription of genes in open chromatin. RNA-seq based transcriptomes of wing imaginal discs over-expressing either CortoCD or RPL12 reveal that both factors deregulate large sets of common genes, which are enriched in heat-response and ribosomal protein genes, suggesting that they could be implicated in dynamic coordination of ribosome biogenesis. Chromatin immunoprecipitation experiments show that Corto and RPL12 bind hsp70 and are similarly recruited on gene body after heat shock. Hence, Corto and RPL12 could be involved together in regulation of gene transcription. We discuss whether pseudo-ribosomal complexes composed of various ribosomal proteins might participate in regulation of gene expression in connection with chromatin regulators.',
'date' => '2012-10-11',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23071455',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '930',
'name' => 'The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts',
'authors' => 'Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J',
'description' => 'Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain and a Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts (MEFs) stably over-expressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the non-coding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2 and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches, showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression and the metabolic control of fibroblasts. ',
'date' => '2012-07-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22825849',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '352',
'name' => 'Mouse IgG SDS US en',
'language' => 'en',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-US-en-GHS_1_0.pdf',
'countries' => 'US',
'modified' => '2020-06-09 15:24:43',
'created' => '2020-06-09 15:24:43',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '350',
'name' => 'Mouse IgG SDS GB en',
'language' => 'en',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-GB-en-GHS_1_0.pdf',
'countries' => 'GB',
'modified' => '2020-06-09 15:23:14',
'created' => '2020-06-09 15:23:14',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '348',
'name' => 'Mouse IgG SDS ES es',
'language' => 'es',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-ES-es-GHS_1_0.pdf',
'countries' => 'ES',
'modified' => '2020-06-09 15:22:03',
'created' => '2020-06-09 15:22:03',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '347',
'name' => 'Mouse IgG SDS DE de',
'language' => 'de',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-DE-de-GHS_1_0.pdf',
'countries' => 'DE',
'modified' => '2020-06-09 15:21:25',
'created' => '2020-06-09 15:21:25',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '351',
'name' => 'Mouse IgG SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-JP-ja-GHS_1_0.pdf',
'countries' => 'JP',
'modified' => '2020-06-09 15:23:45',
'created' => '2020-06-09 15:23:45',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '346',
'name' => 'Mouse IgG SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-BE-nl-GHS_1_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-09 15:20:49',
'created' => '2020-06-09 15:20:49',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '345',
'name' => 'Mouse IgG SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-BE-fr-GHS_1_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-09 15:19:47',
'created' => '2020-06-09 15:19:47',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '349',
'name' => 'Mouse IgG SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-FR-fr-GHS_1_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-09 15:22:38',
'created' => '2020-06-09 15:22:38',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/mouse-igg-15-ug-15-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
)
$pro = array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/bioruptor-pico-sonication-device"><img src="/img/product/shearing_technologies/bioruptor_pico.jpg" alt="Bioruptor pico next gen sequencing " class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">B01060010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Bioruptor® Pico sonication device</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '1862',
'product_id' => '1959',
'related_id' => '1787'
),
'Image' => array(
(int) 0 => array(
'id' => '133',
'name' => 'product/shearing_technologies/bioruptor_pico.jpg',
'alt' => 'Bioruptor pico next gen sequencing ',
'modified' => '2020-11-27 10:00:30',
'created' => '2015-06-10 17:28:55',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1772',
'name' => 'product/shearing_technologies/B010600010.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 15:41:46',
'created' => '2018-02-14 15:41:46',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1773',
'name' => 'product/shearing_technologies/B010600010-2.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 16:18:03',
'created' => '2018-02-14 15:42:00',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1774',
'name' => 'product/shearing_technologies/B01060010-bioruptor-pico-cooler.jpg',
'alt' => 'Pico + cooler',
'modified' => '2018-03-06 11:01:33',
'created' => '2018-03-06 11:01:33',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
'id' => '4167',
'product_id' => '1959',
'application_id' => '43'
)
)
$slugs = array(
(int) 0 => 'chip-qpcr-antibodies'
)
$applications = array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'locale' => 'eng'
)
$description = ''
$name = 'ChIP-qPCR (ab)'
$document = array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '2068',
'product_id' => '1959',
'document_id' => '38'
)
)
$sds = array(
'id' => '349',
'name' => 'Mouse IgG SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-FR-fr-GHS_1_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-09 15:22:38',
'created' => '2020-06-09 15:22:38',
'ProductsSafetySheet' => array(
'id' => '681',
'product_id' => '1959',
'safety_sheet_id' => '349'
)
)
$publication = array(
'id' => '930',
'name' => 'The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts',
'authors' => 'Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J',
'description' => 'Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain and a Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts (MEFs) stably over-expressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the non-coding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2 and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches, showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression and the metabolic control of fibroblasts. ',
'date' => '2012-07-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22825849',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
'id' => '1075',
'product_id' => '1959',
'publication_id' => '930'
)
)
$externalLink = ' <a href="http://www.ncbi.nlm.nih.gov/pubmed/22825849" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: campaign_id [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'product' => array(
'Product' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '328',
'name' => 'Mouse IgG',
'description' => 'The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative contro',
'clonality' => '',
'isotype' => '',
'lot' => '',
'concentration' => '1.0 µg/µl',
'reactivity' => 'none',
'type' => 'Polyclonal',
'purity' => 'Protein A purified ',
'classification' => '',
'application_table' => '',
'storage_conditions' => '',
'storage_buffer' => '',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-02-24 12:10:53',
'created' => '0000-00-00 00:00:00',
'select_label' => '328 - Mouse IgG ( - 1.0 µg/µl - none - Protein A purified - Mouse)'
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/mouse-igg-15-ug-15-ul'
)
$language = 'en'
$meta_keywords = ''
$meta_description = 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.'
$meta_title = 'Mouse IgG (C15400001) | Diagenode'
$product = array(
'Product' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '328',
'name' => 'Mouse IgG',
'description' => 'The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative contro',
'clonality' => '',
'isotype' => '',
'lot' => '',
'concentration' => '1.0 µg/µl',
'reactivity' => 'none',
'type' => 'Polyclonal',
'purity' => 'Protein A purified ',
'classification' => '',
'application_table' => '',
'storage_conditions' => '',
'storage_buffer' => '',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-02-24 12:10:53',
'created' => '0000-00-00 00:00:00',
'select_label' => '328 - Mouse IgG ( - 1.0 µg/µl - none - Protein A purified - Mouse)'
),
'Slave' => array(
(int) 0 => array(
'id' => '211',
'name' => 'C15400001',
'product_id' => '1959',
'modified' => '2017-05-17 15:27:44',
'created' => '2017-05-17 15:27:44'
)
),
'Group' => array(
'Group' => array(
'id' => '211',
'name' => 'C15400001',
'product_id' => '1959',
'modified' => '2017-05-17 15:27:44',
'created' => '2017-05-17 15:27:44'
),
'Master' => array(
'id' => '1959',
'antibody_id' => '328',
'name' => 'Mouse IgG (sample size)',
'description' => '<p><span>The <strong>negative Ctrl</strong> <strong>IgG</strong> from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody. <br /></span></p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-chip.jpg" alt="Mouse IgG Antibody ChIP Grade" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 1. ChIP with the Diagenode mouse IgG negative control antibody</strong><br />ChIP assays were performed using the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) and the “Auto Histone ChIP-seq” kit (cat. No. C01010020) on sheared chromatin from 1 million HeLa cells. Mouse IgG (cat. No. C15400001) was used as a negative IP control. One μg of antibody per ChIP experiment was used for both antibodies. Quantitative PCR was performed with primers specific for the promoters of the active GAPDH and EIF4A2 genes, and for the inactive MYOD1 gene and the Sat2 satellite repeat. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</small></p>
</div>
</div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/C15400001-if.jpg" alt="Mouse IgG Antibody validated in Immunofluorescence" /></p>
</div>
<div class="small-8 columns">
<p><small> <strong>Figure 2. Immunofluorescence with the Diagenode mouse IgG negative control antibody</strong><br />HeLa cells were stained with the Diagenode mouse monoclonal antibody against H3K27ac (cat. No. C15200184) (top) and with DAPI. Mouse IgG (cat. No. C15400001) was used as a negative control (bottom). Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labeled with the H3K27ac or mouse IgG negative control antibody (left) diluted 1:500 in blocking solution followed by an anti-mouse antibody conjugated to Alexa594. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>The negative control IgG from mouse has been extensively validated in chromatin immunoprecipitation (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy mice. This IgG preparation is intended for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. The negative control IgG from mouse should be used in parallel with the specific antibody at the same concentration. It is also included in many of our ChIP and MeDIP kits.</p>',
'label3' => '',
'info3' => '',
'format' => '15 µg/15 µl',
'catalog_number' => 'C15400001-15',
'old_catalog_number' => 'kch-819-015',
'sf_code' => 'C15400001-360',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '75',
'price_USD' => '75',
'price_GBP' => '60',
'price_JPY' => '11750',
'price_CNY' => '',
'price_AUD' => '188',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'mouse-igg-15-ug-15-ul',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2020-11-26 12:46:11',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '7',
'position' => '10',
'parent_id' => '1',
'name' => 'Methylated DNA immunoprecipitation',
'description' => '<div class="row extra-spaced">
<div class="small-12 medium-3 large-3 columns"><center><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank"><img src="https://www.diagenode.com/img/banners/banner-nature-publication-580.png" /></a></center></div>
<div class="small-12 medium-9 large-9 columns">
<h3>Sensitive tumour detection and classification using plasma cell-free DNA methylomes<br /><a href="https://www.ncbi.nlm.nih.gov/pubmed/30429608" target="_blank">Read the publication</a></h3>
<h3 class="c-article-title u-h1" data-test="article-title" itemprop="name headline">Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA<br /><a href="https://www.nature.com/articles/s41596-019-0202-2" target="_blank" title="cfMeDIP-seq Nature Method">Read the method</a></h3>
</div>
</div>
<div class="row">
<div class="large-12 columns"><span>The Methylated DNA Immunoprecipitation is based on the affinity purification of methylated and hydroxymethylated DNA using, respectively, an antibody directed against 5-methylcytosine (5-mC) in the case of MeDIP or 5-hydroxymethylcytosine (5-hmC) in the case of hMeDIP.</span><br />
<h2></h2>
<h2>How it works</h2>
<p>In brief, Methyl DNA IP is performed as follows: Genomic DNA from cultured cells or tissues is prepared, sheared, and then denatured. Then, immunoselection and immunoprecipitation can take place using the antibody directed against 5 methylcytosine and antibody binding beads. After isolation and purification is performed, the IP’d methylated DNA is ready for any subsequent analysis as qPCR, amplification, hybridization on microarrays or next generation sequencing.</p>
<h2>Applications</h2>
<div align="center"><a href="https://www.diagenode.com/en/p/magmedip-kit-x48-48-rxns" class="center alert radius button"> qPCR analysis</a></div>
<div align="center"><a href="https://www.diagenode.com/en/p/magmedip-seq-package-V2-x10" class="center alert radius button"> NGS analysis </a></div>
<h2>Advantages</h2>
<ul style="font-size: 19px;" class="nobullet">
<li><i class="fa fa-arrow-circle-right"></i> <strong>Unaffected</strong> DNA</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>High enrichment</strong> yield</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>Robust</strong> & <strong>reproducible</strong> techniques</li>
<li><i class="fa fa-arrow-circle-right"></i> <strong>NGS</strong> compatible</li>
</ul>
<h2></h2>
</div>
</div>
<div id="gtx-trans" style="position: absolute; left: 17px; top: 652.938px;">
<div class="gtx-trans-icon"></div>
</div>',
'in_footer' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'slug' => 'methylated-dna-immunoprecipitation',
'meta_keywords' => 'Methylated DNA immunoprecipitation,Epigenetic,DNA Methylation,qPCR,5 methylcytosine (5-mC)',
'meta_description' => 'Methylated DNA immunoprecipitation method is based on the affinity purification of methylated DNA using an antibody directed against 5 methylcytosine (5-mC). ',
'meta_title' => 'Methylated DNA immunoprecipitation(MeDIP) - Dna methylation | Diagenode',
'modified' => '2021-08-19 12:08:03',
'created' => '2014-09-14 05:33:34',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '22',
'position' => '40',
'parent_id' => '4',
'name' => 'Isotype controls',
'description' => '<p><span style="font-weight: 400;">Diagenode offers the negative Ctrl IgG from rabbit, rat and mouse. These extensively validated antibodies can be used as negative controls in ChIP, IF, hMeDIP or other experiments performed with specific antibodies made in rabbit, rat or mouse, respectively.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'isotype-controls',
'cookies_tag_id' => null,
'meta_keywords' => 'Isotype controls,DNA immunoprecipitation,Methylated DNA immunoprecipitation',
'meta_description' => 'Diagenode provides Isotype controls for Methylated DNA immunoprecipitation',
'meta_title' => 'Isotype controls for Methylated DNA immunoprecipitation | Diagenode',
'modified' => '2019-07-04 16:19:36',
'created' => '2014-09-30 14:23:34',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '127',
'position' => '10',
'parent_id' => '4',
'name' => 'ChIP-grade antibodies',
'description' => '<div class="row">
<div class="small-10 columns"><center></center>
<p><br />Chromatin immunoprecipitation (<b>ChIP</b>) is a technique to study the associations of proteins with the specific genomic regions in intact cells. One of the most important steps of this protocol is the immunoprecipitation of targeted protein using the antibody specifically recognizing it. The quality of antibodies used in ChIP is essential for the success of the experiment. Diagenode offers extensively validated ChIP-grade antibodies, confirmed for their specificity, and high level of performance in ChIP. Each batch is validated, and batch-specific data are available on the website.</p>
<p></p>
</div>
<div class="small-2 columns"><img src="https://www.diagenode.com/emailing/images/epi-success-guaranteed-icon.png" alt="Epigenetic success guaranteed" /></div>
</div>
<p><strong>ChIP results</strong> obtained with the antibody directed against H3K4me3 (Cat. No. <a href="../p/h3k4me3-polyclonal-antibody-premium-50-ug-50-ul">C15410003</a>). </p>
<div class="row">
<div class="small-12 medium-6 large-6 columns"><img src="https://www.diagenode.com/img/product/antibodies/C15410003-fig1-ChIP.jpg" alt="" width="400" height="315" /> </div>
<div class="small-12 medium-6 large-6 columns">
<p></p>
<p></p>
<p></p>
</div>
</div>
<p></p>
<p>Our aim at Diagenode is to offer the largest collection of highly specific <strong>ChIP-grade antibodies</strong>. We add new antibodies monthly. Find your ChIP-grade antibody in the list below and check more information about tested applications, extensive validation data, and product information.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'chip-grade-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'ChIP-grade antibodies, polyclonal antibody, monoclonal antibody, Diagenode',
'meta_description' => 'Diagenode Offers Extensively Validated ChIP-Grade Antibodies, Confirmed for their Specificity, and high level of Performance in Chromatin Immunoprecipitation ChIP',
'meta_title' => 'Chromatin immunoprecipitation ChIP-grade antibodies | Diagenode',
'modified' => '2021-07-01 10:22:38',
'created' => '2017-02-14 11:16:04',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '726',
'name' => 'Datasheet mouseIgG C15400001',
'description' => 'Datasheet description',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_mouseIgG_C15400001.pdf',
'slug' => 'datasheet-mouseigg-c15400001',
'meta_keywords' => null,
'meta_description' => null,
'modified' => '2015-07-07 11:47:44',
'created' => '2015-07-07 11:47:44',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '250',
'name' => 'product/antibodies/antibody.png',
'alt' => 'Mouse IgG',
'modified' => '2020-11-27 07:00:09',
'created' => '2015-07-17 10:12:18',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4787',
'name' => 'The Effect of Metformin and Carbohydrate-Controlled Diet onDNA Methylation and Gene Expression in the Endometrium of Womenwith Polycystic Ovary Syndrome.',
'authors' => 'Garcia-Gomez E. et al.',
'description' => '<p>Polycystic ovary syndrome (PCOS) is an endocrine disease associated with infertility and metabolic disorders in reproductive-aged women. In this study, we evaluated the expression of eight genes related to endometrial function and their DNA methylation levels in the endometrium of PCOS patients and women without the disease (control group). In addition, eight of the PCOS patients underwent intervention with metformin (1500 mg/day) and a carbohydrate-controlled diet (type and quantity) for three months. Clinical and metabolic parameters were determined, and RT-qPCR and MeDIP-qPCR were used to evaluate gene expression and DNA methylation levels, respectively. Decreased expression levels of , , and genes and increased DNA methylation levels of the promoter were found in the endometrium of PCOS patients compared to controls. After metformin and nutritional intervention, some metabolic and clinical variables improved in PCOS patients. This intervention was associated with increased expression of , , and genes and reduced DNA methylation levels of the promoter in the endometrium of PCOS women. Our preliminary findings suggest that metformin and a carbohydrate-controlled diet improve endometrial function in PCOS patients, partly by modulating DNA methylation of the gene promoter and the expression of genes implicated in endometrial receptivity and insulin signaling.</p>',
'date' => '2023-04-01',
'pmid' => 'https://doi.org/10.3390%2Fijms24076857',
'doi' => '10.3390/ijms24076857',
'modified' => '2023-06-12 08:58:33',
'created' => '2023-05-05 12:34:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4127',
'name' => 'The histone modification H3K4me3 is altered at the locus in Alzheimer'sdisease brain.',
'authors' => 'Smith, Adam et al.',
'description' => '<p>Several epigenome-wide association studies of DNA methylation have highlighted altered DNA methylation in the gene in Alzheimer's disease (AD) brain samples. However, no study has specifically examined histone modifications in the disease. We use chromatin immunoprecipitation-qPCR to quantify tri-methylation at histone 3 lysine 4 (H3K4me3) and 27 (H3K27me3) in the gene in entorhinal cortex from donors with high (n = 59) or low (n = 29) Alzheimer's disease pathology. We demonstrate decreased levels of H3K4me3, a marker of active gene transcription, with no change in H3K27me3, a marker of inactive genes. H3K4me3 is negatively correlated with DNA methylation in specific regions of the gene. Our study suggests that the gene shows altered epigenetic marks indicative of reduced gene activation in Alzheimer's disease.</p>',
'date' => '2021-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33815817',
'doi' => '10.2144/fsoa-2020-0161',
'modified' => '2021-12-07 10:16:08',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4188',
'name' => 'Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage.',
'authors' => 'Ait-Ammar A. et al.',
'description' => '<p>HIV-1 latency generates reservoirs that prevent viral eradication by the current therapies. To find strategies toward an HIV cure, detailed understandings of the molecular mechanisms underlying establishment and persistence of the reservoirs are needed. The cellular transcription factor KAP1 is known as a potent repressor of gene transcription. Here we report that KAP1 represses HIV-1 gene expression in myeloid cells including microglial cells, the major reservoir of the central nervous system. Mechanistically, KAP1 interacts and colocalizes with the viral transactivator Tat to promote its degradation via the proteasome pathway and repress HIV-1 gene expression. In myeloid models of latent HIV-1 infection, the depletion of KAP1 increased viral gene elongation and reactivated HIV-1 expression. Bound to the latent HIV-1 promoter, KAP1 associates and cooperates with CTIP2, a key epigenetic silencer of HIV-1 expression in microglial cells. In addition, Tat and CTIP2 compete for KAP1 binding suggesting a dynamic modulation of the KAP1 cellular partners upon HIV-1 infection. Altogether, our results suggest that KAP1 contributes to the establishment and the persistence of HIV-1 latency in myeloid cells.</p>',
'date' => '2021-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33514850',
'doi' => '10.1038/s41598-021-82164-w',
'modified' => '2022-01-05 15:08:41',
'created' => '2021-12-06 15:53:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4082',
'name' => 'p53 directly represses human LINE1 transposons.',
'authors' => 'Tiwari, Bhavana and Jones, Amanda E and Caillet, Candace J and Das, Simantiand Royer, Stephanie K and Abrams, John M',
'description' => '<p>p53 is a potent tumor suppressor and commonly mutated in human cancers. Recently, we demonstrated that p53 genes act to restrict retrotransposons in germline tissues of flies and fish but whether this activity is conserved in somatic human cells is not known. Here we show that p53 constitutively restrains human LINE1s by cooperatively engaging sites in the 5'UTR and stimulating local deposition of repressive histone marks at these transposons. Consistent with this, the elimination of p53 or the removal of corresponding binding sites in LINE1s, prompted these retroelements to become hyperactive. Concurrently, p53 loss instigated chromosomal rearrangements linked to LINE sequences and also provoked inflammatory programs that were dependent on reverse transcriptase produced from LINE1s. Taken together, our observations establish that p53 continuously operates at the LINE1 promoter to restrict autonomous copies of these mobile elements in human cells. Our results further suggest that constitutive restriction of these retroelements may help to explain tumor suppression encoded by p53, since erupting LINE1s produced acute oncogenic threats when p53 was absent.</p>',
'date' => '2020-10-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/33060137',
'doi' => '10.1101/gad.343186.120',
'modified' => '2021-03-15 16:59:03',
'created' => '2021-02-18 10:21:53',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '3953',
'name' => 'Attenuated Epigenetic Suppression of Muscle Stem Cell Necroptosis Is Required for Efficient Regeneration of Dystrophic Muscles.',
'authors' => 'Sreenivasan K, Ianni A, Künne C, Strilic B, Günther S, Perdiguero E, Krüger M, Spuler S, Offermanns S, Gómez-Del Arco P, Redondo JM, Munoz-Canoves P, Kim J, Braun T',
'description' => '<p>Somatic stem cells expand massively during tissue regeneration, which might require control of cell fitness, allowing elimination of non-competitive, potentially harmful cells. How or if such cells are removed to restore organ function is not fully understood. Here, we show that a substantial fraction of muscle stem cells (MuSCs) undergo necroptosis because of epigenetic rewiring during chronic skeletal muscle regeneration, which is required for efficient regeneration of dystrophic muscles. Inhibition of necroptosis strongly enhances suppression of MuSC expansion in a non-cell-autonomous manner. Prevention of necroptosis in MuSCs of healthy muscles is mediated by the chromatin remodeler CHD4, which directly represses the necroptotic effector Ripk3, while CHD4-dependent Ripk3 repression is dramatically attenuated in dystrophic muscles. Loss of Ripk3 repression by inactivation of Chd4 causes massive necroptosis of MuSCs, abolishing regeneration. Our study demonstrates how programmed cell death in MuSCs is tightly controlled to achieve optimal tissue regeneration.</p>',
'date' => '2020-05-19',
'pmid' => 'http://www.pubmed.gov/32433961',
'doi' => '10.1016/j.celrep.2020.107652',
'modified' => '2020-08-17 09:51:58',
'created' => '2020-08-10 12:12:25',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '3834',
'name' => 'Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite.',
'authors' => 'Liu J, An Z, Luo J, Li J, Li F, Zhang Z',
'description' => '<p>MOTIVATION: RNA 5-methylcytosine (m5C) is a type of post-transcriptional modification that may be involved in numerous biological processes and tumorigenesis. RNA m5C can be profiled at single-nucleotide resolution by high-throughput sequencing of RNA treated with bisulfite (RNA-BisSeq). However, the exploration of transcriptome-wide profile and potential function of m5C in splicing remains to be elucidated due to lack of isoform level m5C quantification tool. RESULTS: We developed a computational package to quantify Epitranscriptomal RNA m5C at the transcript isoform level (named Episo). Episo consists of three tools, mapper, quant and Bisulfitefq, for mapping, quantifying, and simulating RNA-BisSeq data, respectively. The high accuracy of Episo was validated using an improved m5C-specific methylated RNA immunoprecipitation (meRIP) protocol, as well as a set of in silico experiments. By applying Episo to public human and mouse RNA-BisSeq data, we found that the RNA m5C is not evenly distributed among the transcript isoforms, implying the m5C may subject to be regulated at isoform level. AVAILABILITY: Episo is released under the GNU GPLv3+ license. The resource code Episo is freely accessible from https://github.com/liujunfengtop/Episo (with Tophat/cufflink) and https://github.com/liujunfengtop/Episo/tree/master/Episo_Kallisto (with Kallisto). SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.</p>',
'date' => '2019-12-03',
'pmid' => 'http://www.pubmed.gov/31794005',
'doi' => '10.1093/bioinformatics/btz900/5651015',
'modified' => '2020-02-25 13:26:22',
'created' => '2020-02-13 10:02:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '3499',
'name' => 'Centromeres License the Mitotic Condensation of Yeast Chromosome Arms.',
'authors' => 'Kruitwagen T, Chymkowitch P, Denoth-Lippuner A, Enserink J, Barral Y',
'description' => '<p>During mitosis, chromatin condensation shapes chromosomes as separate, rigid, and compact sister chromatids to facilitate their segregation. Here, we show that, unlike wild-type yeast chromosomes, non-chromosomal DNA circles and chromosomes lacking a centromere fail to condense during mitosis. The centromere promotes chromosome condensation strictly in cis through recruiting the kinases Aurora B and Bub1, which trigger the autonomous condensation of the entire chromosome. Shugoshin and the deacetylase Hst2 facilitated spreading the condensation signal to the chromosome arms. Targeting Aurora B to DNA circles or centromere-ablated chromosomes or releasing Shugoshin from PP2A-dependent inhibition bypassed the centromere requirement for condensation and enhanced the mitotic stability of DNA circles. Our data indicate that yeast cells license the chromosome-autonomous condensation of their chromatin in a centromere-dependent manner, excluding from this process non-centromeric DNA and thereby inhibiting their propagation.</p>',
'date' => '2018-10-18',
'pmid' => 'http://www.pubmed.org/30318142',
'doi' => '10.1016/j.cell.2018.09.012',
'modified' => '2019-02-27 15:44:25',
'created' => '2019-02-27 12:54:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '3235',
'name' => 'A novel FOXA1/ESR1 interacting pathway: A study of Oncomine™ breast cancer microarrays',
'authors' => 'Chaudhary S. et al.',
'description' => '<p>Forkhead box protein A1 (FOXA1) is essential for the growth and differentiation of breast epithelium, and has a favorable outcome in breast cancer (BC). Elevated <i>FOXA1</i> expression in BC also facilitates hormone responsiveness in estrogen receptor (<i>ESR</i>)-positive BC. However, the interaction between these two pathways is not fully understood. <i>FOXA1</i> and GATA binding protein 3 (<i>GATA3</i>) along with <i>ESR1</i> expression are responsible for maintaining a luminal phenotype, thus suggesting the existence of a strong association between them. The present study utilized the Oncomine™ microarray database to identify <i>FOXA1:ESR1</i> and <i>FOXA1:ESR1:GATA3</i> co-expression co-regulated genes. Oncomine™ analysis revealed 115 and 79 overlapping genes clusters in <i>FOXA1:ESR1</i> and <i>FOXA1:ESR1:GATA3</i> microarrays, respectively. Five ESR1 direct target genes [trefoil factor 1 (<i>TFF1/PS2</i>), <i>B-cell lymphoma</i> 2 (<i>BCL2</i>), seven in absentia homolog 2 (<i>SIAH2</i>), cellular myeloblastosis viral oncogene homolog (<i>CMYB</i>) and progesterone receptor (<i>PGR</i>)] were detected in the co-expression clusters. To further investigate the role of FOXA1 in ESR1-positive cells, MCF7 cells were transfected with a <i>FOXA1</i> expression plasmid, and it was observed that the direct target genes of ESR1 (<i>PS2, BCL2, SIAH2</i> and <i>PGR</i>) were significantly regulated upon transfection. Analysis of one of these target genes, <i>PS2</i>, revealed the presence of two FOXA1 binding sites in the vicinity of the estrogen response element (ERE), which was confirmed by binding assays. Under estrogen stimulation, FOXA1 protein was recruited to the FOXA1 site and could also bind to the ERE site (although in minimal amounts) in the <i>PS2</i> promoter. Co-transfection of <i>FOXA1</i>/<i>ESR1</i> expression plasmids demonstrated a significantly regulation of the target genes identified in the <i>FOXA1</i>/<i>ESR1</i> multi-arrays compared with only <i>FOXA1</i> transfection, which was suggestive of a synergistic effect of <i>ESR1</i> and <i>FOXA1</i> on the target genes. In summary, the present study identified novel <i>FOXA1</i>, <i>ESR1</i> and <i>GATA</i>3 co-expressed genes that may be involved in breast tumorigenesis.</p>',
'date' => '2017-08-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28789340',
'doi' => '',
'modified' => '2017-08-28 09:30:38',
'created' => '2017-08-28 09:30:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '3236',
'name' => 'TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors',
'authors' => 'Ancey P.B. et al.',
'description' => '<p>Understanding the processes that govern liver progenitor cell differentiation has important implications for the design of strategies targeting chronic liver diseases, whereby regeneration of liver tissue is critical. Although DNA methylation (5mC) and hydroxymethylation (5hmC) are highly dynamic during early embryonic development, less is known about their roles at later stages of differentiation. Using an in vitro model of hepatocyte differentiation, we show here that 5hmC precedes the expression of promoter 1 (P1)-dependent isoforms of HNF4A, a master transcription factor of hepatocyte identity. 5hmC and HNF4A expression from P1 are dependent on ten-eleven translocation (TET) dioxygenases. In turn, the liver pioneer factor FOXA2 is necessary for TET1 binding to the P1 locus. Both FOXA2 and TETs are required for the 5hmC-related switch in HNF4A expression. The epigenetic event identified here may be a key step for the establishment of the hepatocyte program by HNF4A.</p>',
'date' => '2017-07-11',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28648900',
'doi' => '',
'modified' => '2017-08-28 10:24:16',
'created' => '2017-08-28 09:44:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '3113',
'name' => 'Differentiation of Mouse Enteric Nervous System Progenitor Cells is Controlled by Endothelin 3 and Requires Regulation of Ednrb by SOX10 and ZEB2',
'authors' => 'Watanabe Y. et al.',
'description' => '<h2 class="sectionTitle"></h2>
<h3 class="sectionTitle">Background & Aims</h3>
<p>Maintenance and differentiation of progenitor cells in the developing enteric nervous system (ENS) are controlled by molecules such as the signaling protein endothelin 3 (EDN3), its receptor (the endothelin receptor type B, EDNRB), and the transcription factors SRY-box 10 (SOX10) and zinc finger E-box binding homeobox 2 (ZEB2). We used enteric progenitor cell (EPC) cultures and mice to study the roles of these proteins in enteric neurogenesis and their cross regulation.</p>
<h3 class="sectionTitle">Methods</h3>
<p>We performed studies in mice with a <em>Zeb2</em> loss-of-function mutation (<em>Zeb2</em><sup>Δ</sup>) and mice carrying a spontaneous recessive mutation that prevents conversion EDN3 to its active form (<em>Edn3</em><sup><em>ls</em></sup>). EPC cultures issued from embryos that expressed only wild-type <em>Zeb2</em> (<em>Zeb2</em><sup>+/+</sup> EPCs) or were heterozygous for the mutation (<em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs) were exposed to EDN3; we analyzed the effects on cell differentiation using immunocytochemistry. In parallel, <em>Edn3</em><sup><em>ls</em></sup> mice were crossed with <em>Zeb2</em><sup><em>Δ/+</em></sup>mice; intestinal tissues were collected from embryos for immunohistochemical analyses. We investigated regulation of the <em>EDNRB</em> gene in transactivation and chromatin immunoprecipitation assays; results were validated in functional rescue experiments using transgenes expression in EPCs from retroviral vectors.</p>
<h3 class="sectionTitle">Results</h3>
<p><em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs had increased neuronal differentiation compared to <em>Zeb2</em><sup><em>+/+</em></sup> cells. When exposed to EDN3, <em>Zeb2</em><sup>+/+</sup> EPCs continued expression of ZEB2 but did not undergo any neuronal differentiation. Incubation of <em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs with EDN3, on the other hand, resulted in only partial inhibition of neuronal differentiation. This indicated that 2 copies of <em>Zeb2</em> are required for EDN3 to prevent neuronal differentiation. Mice with combined mutations in <em>Zeb2</em> and <em>Edn3</em> (double mutants) had more severe enteric anomalies and increased neuronal differentiation compared to mice with mutations in either gene alone. The transcription factors SOX10 and ZEB2 directly activated the <em>EDNRB</em> promoter. Overexpression of EDNRB in <em>Zeb2</em><sup><em>Δ/+</em></sup> EPCs restored inhibition of neuronal differentiation, similar to incubation of <em>Zeb2</em><sup>+/+</sup> EPCs with EDN3.</p>
<h3 class="sectionTitle">Conclusions</h3>
<p>In studies of cultured EPCs and mice, we found that control of differentiation of mouse enteric nervous system progenitor cells by EDN3 requires regulation of <em>Ednrb</em> expression by SOX10 and ZEB2.</p>',
'date' => '2017-01-05',
'pmid' => 'http://www.gastrojournal.org/article/S0016-5085(17)30002-1/abstract?referrer=http%3A%2F%2Fwww.gastrojournal.org%2Farticle%2FS0016-5085%2817%2930002-1%2Ffulltext',
'doi' => '',
'modified' => '2017-01-06 10:04:00',
'created' => '2017-01-06 10:04:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '3183',
'name' => 'High resolution methylation analysis of the HoxA5 regulatory region in different somatic tissues of laboratory mouse during development',
'authors' => 'Sinha P. et al.',
'description' => '<p>Homeobox genes encode a group of DNA binding regulatory proteins whose key function occurs in the spatial-temporal organization of genome during embryonic development and differentiation. The role of these Hox genes during ontogenesis makes it an important model for research. HoxA5 is a member of Hox gene family playing a central role during axial body patterning and morphogenesis. DNA modification studies have shown that the function of Hox genes is partly governed by the methylation-mediated gene expression regulation. Therefore the study aimed to investigate the role of epigenetic events in regulation of tissue-specific expression pattern of HoxA5 gene during mammalian development. The methodology adopted were sodium bisulfite genomic DNA sequencing, quantitative real-time PCR and chromatin-immunoprecipitation (ChIP). Methylation profiling of HoxA5 gene promoter shows higher methylation in adult as compared to fetus in various somatic tissues of mouse being highest in adult spleen. However q-PCR results show higher expression during fetal stages being highest in fetal intestine followed by brain, liver and spleen. These results clearly indicate a strict correlation between DNA methylation and tissue-specific gene expression. The findings of chromatin-immunoprecipitation (ChIP) have also reinforced that epigenetic event like DNA methylation plays important role in the regulation of tissue specific expression of HoxA5.</p>',
'date' => '2017-01-02',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28363633',
'doi' => '',
'modified' => '2017-05-22 09:48:38',
'created' => '2017-05-22 09:48:38',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '3054',
'name' => 'Overexpression of histone demethylase Fbxl10 leads to enhanced migration in mouse embryonic fibroblasts.',
'authors' => 'Rohde M. et al.',
'description' => '<p>Cell migration is a central process in the development and maintenance of multicellular organisms. Tissue formation during embryonic development, wound healing, immune responses and invasive tumors all require the orchestrated movement of cells to specific locations. Histone demethylase proteins alter transcription by regulating the chromatin state at specific gene loci. FBXL10 is a conserved and ubiquitously expressed member of the JmjC domain-containing histone demethylase family and is implicated in the demethylation of H3K4me3 and H3K36me2 and thereby removing active chromatin marks. However, the physiological role of FBXL10 in vivo remains largely unknown. Therefore, we established an inducible gain of function model to analyze the role of Fbxl10 and compared wild-type with Fbxl10 overexpressing mouse embryonic fibroblasts (MEFs). Our study shows that overexpression of Fbxl10 in MEFs doesn't influence the proliferation capability but leads to an enhanced migration capacity in comparison to wild-type MEFs. Transcriptome and ChIP-seq experiments demonstrated that Fbxl10 binds to genes involved in migration like Areg, Mdk, Lmnb1, Thbs1, Mgp and Cxcl12. Taken together, our results strongly suggest that Fbxl10 plays a critical role in migration by binding to the promoter region of migration-associated genes and thereby might influences cell behaviour to a possibly more aggressive phenotype.</p>',
'date' => '2016-09-17',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27646113',
'doi' => '',
'modified' => '2016-10-24 14:35:45',
'created' => '2016-10-24 14:35:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '2979',
'name' => 'Methylation of the Sox9 and Oct4 promoters and its correlation with gene expression during testicular development in the laboratory mouse',
'authors' => 'Pamnani M et al.',
'description' => '<p>Sox9 and Oct4 are two important regulatory factors involved in mammalian development. Sox9, a member of the group E Sox transcription factor family, has a crucial role in the development of the genitourinary system, while Oct4, commonly known as octamer binding transcription factor 4, belongs to class V of the transcription family. The expression of these two proteins exhibits a dynamic pattern with regard to their expression sites and levels. The aim of this study was to investigate the role of de novo methylation in the regulation of the tissue- and site-specific expression of these proteins. The dynamics of the de novo methylation of 15 CpGs and six CpGs in Sox9 and Oct4 respectively, was studied with sodium bisulfite genomic DNA sequencing in mouse testis at different developmental stages. Consistent methylation of three CpGs was observed in adult ovary in which the expression of Sox9 was feeble, while the level of methylation in somatic tissue was greater in Oct4 compared to germinal tissue. The promoter-chromatin status of Sox9 was also studied with a chromatin immune-precipitation assay.</p>',
'date' => '2016-07-04',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27381637',
'doi' => '10.1590/1678-4685-GMB-2015-0172',
'modified' => '2016-07-11 12:31:08',
'created' => '2016-07-11 12:31:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '2897',
'name' => 'Overexpression of caspase 7 is ERα dependent to affect proliferation and cell growth in breast cancer cells by targeting p21(Cip)',
'authors' => 'Chaudhary S, Madhukrishna B, Adhya AK, Keshari S, Mishra SK',
'description' => '<p>Caspase 7 (CASP7) expression has important function during cell cycle progression and cell growth in certain cancer cells and is also involved in the development and differentiation of dental tissues. However, the function of CASP7 in breast cancer cells is unclear. The aim of this study was to analyze the expression of CASP7 in breast carcinoma patients and determine the role of CASP7 in regulating tumorigenicity in breast cancer cells. In this study, we show that the CASP7 expression is high in breast carcinoma tissues compared with normal counterpart. The ectopic expression of CASP7 is significantly associated with ERα expression status and persistently elevated in different stages of the breast tumor grades. High level of CASP7 expression showed better prognosis in breast cancer patients with systemic endocrine therapy as observed from Kaplan-Meier analysis. S3 and S4, estrogen responsive element (ERE) in the CASP7 promoter, is important for estrogen-ERα-mediated CASP7 overexpression. Increased recruitment of p300, acetylated H3 and pol II in the ERE region of CASP7 promoter is observed after hormone stimulation. Ectopic expression of CASP7 in breast cancer cells results in cell growth and proliferation inhibition via p21(Cip) reduction, whereas small interfering RNA (siRNA) mediated reduction of CASP7 rescued p21(Cip) levels. We also show that pro- and active forms of CASP7 is located in the nucleus apart from cytoplasmic region of breast cancer cells. The proliferation and growth of breast cancer cells is significantly reduced by broad-spectrum peptide inhibitors and siRNA of CASP7. Taken together, our findings show that CASP7 is aberrantly expressed in breast cancer and contributes to cell growth and proliferation by downregulating p21(Cip) protein, suggesting that targeting CASP7-positive breast cancer could be one of the potential therapeutic strategies.</p>',
'date' => '2016-04-18',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/27089142',
'doi' => '10.1038/oncsis.2016.12',
'modified' => '2016-04-28 10:15:00',
'created' => '2016-04-28 10:15:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '2834',
'name' => 'Methylated DNA Immunoprecipitation Analysis of Mammalian Endogenous Retroviruses.',
'authors' => 'Rebollo R, Mager DL',
'description' => '<p>Endogenous retroviruses are repetitive sequences found abundantly in mammalian genomes which are capable of modulating host gene expression. Nevertheless, most endogenous retrovirus copies are under tight epigenetic control via histone-repressive modifications and DNA methylation. Here we describe a common method used in our laboratory to detect, quantify, and compare mammalian endogenous retrovirus DNA methylation. More specifically we describe methylated DNA immunoprecipitation (MeDIP) followed by quantitative PCR.</p>',
'date' => '2016-02-20',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26895065',
'doi' => '10.1007/978-1-4939-3372-3_23',
'modified' => '2016-03-02 10:37:15',
'created' => '2016-03-02 10:22:16',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '2842',
'name' => 'Chromatin Immunoprecipitation Assay for the Identification of Arabidopsis Protein-DNA Interactions In Vivo',
'authors' => 'Komar DN, Mouriz A, Jarillo JA, Piñeiro M',
'description' => '<p>Intricate gene regulatory networks orchestrate biological processes and developmental transitions in plants. Selective transcriptional activation and silencing of genes mediate the response of plants to environmental signals and developmental cues. Therefore, insights into the mechanisms that control plant gene expression are essential to gain a deep understanding of how biological processes are regulated in plants. The chromatin immunoprecipitation (ChIP) technique described here is a procedure to identify the DNA-binding sites of proteins in genes or genomic regions of the model species Arabidopsis thaliana. The interactions with DNA of proteins of interest such as transcription factors, chromatin proteins or posttranslationally modified versions of histones can be efficiently analyzed with the ChIP protocol. This method is based on the fixation of protein-DNA interactions in vivo, random fragmentation of chromatin, immunoprecipitation of protein-DNA complexes with specific antibodies, and quantification of the DNA associated with the protein of interest by PCR techniques. The use of this methodology in Arabidopsis has contributed significantly to unveil transcriptional regulatory mechanisms that control a variety of plant biological processes. This approach allowed the identification of the binding sites of the Arabidopsis chromatin protein EBS to regulatory regions of the master gene of flowering FT. The impact of this protein in the accumulation of particular histone marks in the genomic region of FT was also revealed through ChIP analysis.</p>',
'date' => '2016-01-14',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26863263',
'doi' => '10.3791/53422',
'modified' => '2017-01-04 14:16:52',
'created' => '2016-03-09 17:05:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '2801',
'name' => 'The genetic association of RUNX3 with ankylosing spondylitis can be explained by allele-specific effects on IRF4 recruitment that alter gene expression',
'authors' => 'Matteo Vecellio, Amity R Roberts, Carla J Cohen, Adrian Cortes, Julian C Knight, Paul Bowness, B Paul Wordsworth',
'description' => '<p>The authors sought to identify the functional basis for the genetic association of single nucleotide polymorphisms (SNP), upstream of the RUNX3 promoter, with ankylosing spondylitis (AS). They performed conditional analysis of genetic association data and used ENCODE data on chromatin remodelling and transcription factor (TF) binding sites to identify the primary AS-associated regulatory SNP in the RUNX3 region. The functional effects of this SNP were tested in luciferase reporter assays. Its effects on TF binding were investigated by electrophoretic mobility gel shift assays and chromatin immunoprecipitation. RUNX3 mRNA levels were compared in primary CD8+ T cells of AS risk and protective genotypes by real-time PCR. They identified a regulatory region upstream of RUNX3 that is modulated byrs4648889. The risk allele decreases TF binding (including IRF4) and reduces reporter activity and RUNX3 expression. These findings may have important implications for understanding the role of T cells and other immune cells in AS.</p>',
'date' => '2015-10-09',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26452539',
'doi' => '10.1136/annrheumdis-2015-207490',
'modified' => '2016-04-25 09:56:04',
'created' => '2015-12-07 06:37:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '2852',
'name' => 'ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells',
'authors' => 'Al-Furoukh N, Ianni A, Nolte H, Hölper S, Krüger M, Wanrooij S, Braun T',
'description' => '<p>Proteostasis is crucial for life and maintained by cellular chaperones and proteases. One major mitochondrial protease is the ClpXP complex, which is comprised of a catalytic ClpX subunit and a proteolytic ClpP subunit. Based on two separate observations, we hypothesized that ClpX may play a leading role in the cellular function of ClpXP. Therefore, we analyzed the effect of ClpX overexpression on a myoblast proteome by quantitative proteomics. ClpX overexpression results in the upregulation of markers of the mitochondrial proteostasis pathway, known as the "mitochondrial unfolded protein response" (UPRmt). Although this pathway is described in detail in Caenorhabditis elegans, it is not clear whether it is conserved in mammals. Therefore, we compared features of the classical nematode UPRmt with our mammalian ClpX-triggered UPRmt dataset. We show that they share the same retrograde mitochondria-to-nucleus signaling pathway that involves the key UPRmt transcription factor CHOP (also known as Ddit3, CEBPZ or GADD153). In conclusion, our data confirm the existence of a mammalian UPRmt that has great similarity to the C. elegans pathway. Furthermore, our results illustrate that ClpX overexpression is a good and simple model to study the underlying mechanisms of the UPRmt in mammalian cells.</p>',
'date' => '2015-10-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26142927',
'doi' => '10.1016/j.bbamcr.2015.06.016',
'modified' => '2016-03-14 10:20:09',
'created' => '2016-03-14 10:20:09',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '2890',
'name' => 'Endothelial Msx1 transduces hemodynamic changes into an arteriogenic remodeling response.',
'authors' => 'Vandersmissen I, Craps S, Depypere M, Coppiello G, van Gastel N, Maes F, Carmeliet G, Schrooten J, Jones EA, Umans L, Devlieger R, Koole M, Gheysens O, Zwijsen A, Aranguren XL, Luttun A',
'description' => '<p>Collateral remodeling is critical for blood flow restoration in peripheral arterial disease and is triggered by increasing fluid shear stress in preexisting collateral arteries. So far, no arterial-specific mediators of this mechanotransduction response have been identified. We show that muscle segment homeobox 1 (MSX1) acts exclusively in collateral arterial endothelium to transduce the extrinsic shear stimulus into an arteriogenic remodeling response. MSX1 was specifically up-regulated in remodeling collateral arteries. MSX1 induction in collateral endothelial cells (ECs) was shear stress driven and downstream of canonical bone morphogenetic protein-SMAD signaling. Flow recovery and collateral remodeling were significantly blunted in EC-specific Msx1/2 knockout mice. Mechanistically, MSX1 linked the arterial shear stimulus to arteriogenic remodeling by activating the endothelial but not medial layer to a proinflammatory state because EC but not smooth muscle cellMsx1/2 knockout mice had reduced leukocyte recruitment to remodeling collateral arteries. This reduced leukocyte infiltration in EC Msx1/2 knockout mice originated from decreased levels of intercellular adhesion molecule 1 (ICAM1)/vascular cell adhesion molecule 1 (VCAM1), whose expression was also in vitro driven by promoter binding of MSX1.</p>',
'date' => '2015-09-28',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26391659',
'doi' => ' 10.1083/jcb.201502003',
'modified' => '2016-04-12 10:44:22',
'created' => '2016-04-12 10:44:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '2846',
'name' => 'DNA methylation directs functional maturation of pancreatic β cells',
'authors' => 'Dhawan S, Tschen SI, Zeng C, Guo T, Hebrok M, Matveyenko A, Bhushan A.',
'description' => '<p>Pancreatic β cells secrete insulin in response to postprandial increases in glucose levels to prevent hyperglycemia and inhibit insulin secretion under fasting conditions to protect against hypoglycemia. β cells lack this functional capability at birth and acquire glucose-stimulated insulin secretion (GSIS) during neonatal life. Here, we have shown that during postnatal life, the de novo DNA methyltransferase DNMT3A initiates a metabolic program by repressing key genes, thereby enabling the coupling of insulin secretion to glucose levels. In a murine model, β cell-specific deletion of Dnmt3a prevented the metabolic switch, resulting in loss of GSIS. DNMT3A bound to the promoters of the genes encoding hexokinase 1 (HK1) and lactate dehydrogenase A (LDHA) - both of which regulate the metabolic switch - and knockdown of these two key DNMT3A targets restored the GSIS response in islets from animals with β cell-specific Dnmt3a deletion. Furthermore, DNA methylation-mediated repression of glucose-secretion decoupling genes to modulate GSIS was conserved in human β cells. Together, our results reveal a role for DNA methylation to direct the acquisition of pancreatic β cell function.</p>',
'date' => '2015-07-01',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26098213',
'doi' => '10.1172/JCI79956',
'modified' => '2016-03-11 14:16:44',
'created' => '2016-03-11 14:16:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '2049',
'name' => 'Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions.',
'authors' => 'Tapias A, Zhou ZW, Shi Y, Chong Z, Wang P, Groth M, Platzer M, Huttner W, Herceg Z, Yang YG, Wang ZQ',
'description' => 'Fate decisions in neural progenitor cells are orchestrated via multiple pathways, and the role of histone acetylation in these decisions has been ascribed to a general function promoting gene activation. Here, we show that the histone acetyltransferase (HAT) cofactor transformation/transcription domain-associated protein (Trrap) specifically regulates activation of cell-cycle genes, thereby integrating discrete cell-intrinsic programs of cell-cycle progression and epigenetic regulation of gene transcription in order to control neurogenesis. Deletion of Trrap impairs recruitment of HATs and transcriptional machinery specifically to E2F cell-cycle target genes, disrupting their transcription with consequent cell-cycle lengthening specifically within cortical apical neural progenitors (APs). Consistently, Trrap conditional mutants exhibit microcephaly because of premature differentiation of APs into intermediate basal progenitors and neurons, and overexpressing cell-cycle regulators in vivo can rescue these premature differentiation defects. These results demonstrate an essential and highly specific role for Trrap-mediated histone regulation in controlling cell-cycle progression and neurogenesis.',
'date' => '2014-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/24792116',
'doi' => '',
'modified' => '2015-07-24 15:39:02',
'created' => '2015-07-24 15:39:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '1493',
'name' => 'Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks.',
'authors' => 'Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gäbel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D',
'description' => 'The chromosome 9p21 (Chr9p21) locus of coronary artery disease has been identified in the first surge of genome-wide association and is the strongest genetic factor of atherosclerosis known today. Chr9p21 encodes the long non-coding RNA (ncRNA) antisense non-coding RNA in the INK4 locus (ANRIL). ANRIL expression is associated with the Chr9p21 genotype and correlated with atherosclerosis severity. Here, we report on the molecular mechanisms through which ANRIL regulates target-genes in trans, leading to increased cell proliferation, increased cell adhesion and decreased apoptosis, which are all essential mechanisms of atherogenesis. Importantly, trans-regulation was dependent on Alu motifs, which marked the promoters of ANRIL target genes and were mirrored in ANRIL RNA transcripts. ANRIL bound Polycomb group proteins that were highly enriched in the proximity of Alu motifs across the genome and were recruited to promoters of target genes upon ANRIL over-expression. The functional relevance of Alu motifs in ANRIL was confirmed by deletion and mutagenesis, reversing trans-regulation and atherogenic cell functions. ANRIL-regulated networks were confirmed in 2280 individuals with and without coronary artery disease and functionally validated in primary cells from patients carrying the Chr9p21 risk allele. Our study provides a molecular mechanism for pro-atherogenic effects of ANRIL at Chr9p21 and suggests a novel role for Alu elements in epigenetic gene regulation by long ncRNAs.',
'date' => '2013-07-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23861667',
'doi' => '',
'modified' => '2015-07-24 15:39:00',
'created' => '2015-07-24 15:39:00',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '1285',
'name' => 'Genome-wide analysis of LXRα activation reveals new transcriptional networks in human atherosclerotic foam cells.',
'authors' => 'Feldmann R, Fischer C, Kodelja V, Behrens S, Haas S, Vingron M, Timmermann B, Geikowski A, Sauer S',
'description' => 'Increased physiological levels of oxysterols are major risk factors for developing atherosclerosis and cardiovascular disease. Lipid-loaded macrophages, termed foam cells, are important during the early development of atherosclerotic plaques. To pursue the hypothesis that ligand-based modulation of the nuclear receptor LXRα is crucial for cell homeostasis during atherosclerotic processes, we analysed genome-wide the action of LXRα in foam cells and macrophages. By integrating chromatin immunoprecipitation-sequencing (ChIP-seq) and gene expression profile analyses, we generated a highly stringent set of 186 LXRα target genes. Treatment with the nanomolar-binding ligand T0901317 and subsequent auto-regulatory LXRα activation resulted in sequence-dependent sharpening of the genome-binding patterns of LXRα. LXRα-binding loci that correlated with differential gene expression revealed 32 novel target genes with potential beneficial effects, which in part explained the implications of disease-associated genetic variation data. These observations identified highly integrated LXRα ligand-dependent transcriptional networks, including the APOE/C1/C4/C2-gene cluster, which contribute to the reversal of cholesterol efflux and the dampening of inflammation processes in foam cells to prevent atherogenesis.',
'date' => '2013-04-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23393188',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '1078',
'name' => 'New partners in regulation of gene expression: the enhancer of trithorax and polycomb corto interacts with methylated ribosomal protein l12 via its chromodomain.',
'authors' => 'Coléno-Costes A, Jang SM, de Vanssay A, Rougeot J, Bouceba T, Randsholt NB, Gibert JM, Le Crom S, Mouchel-Vielh E, Bloyer S, Peronnet F',
'description' => 'Chromodomains are found in many regulators of chromatin structure, and most of them recognize methylated lysines on histones. Here, we investigate the role of the Drosophila melanogaster protein Corto's chromodomain. The Enhancer of Trithorax and Polycomb Corto is involved in both silencing and activation of gene expression. Over-expression of the Corto chromodomain (CortoCD) in transgenic flies shows that it is a chromatin-targeting module, critical for Corto function. Unexpectedly, mass spectrometry analysis reveals that polypeptides pulled down by CortoCD from nuclear extracts correspond to ribosomal proteins. Furthermore, real-time interaction analyses demonstrate that CortoCD binds with high affinity RPL12 tri-methylated on lysine 3. Corto and RPL12 co-localize with active epigenetic marks on polytene chromosomes, suggesting that both are involved in fine-tuning transcription of genes in open chromatin. RNA-seq based transcriptomes of wing imaginal discs over-expressing either CortoCD or RPL12 reveal that both factors deregulate large sets of common genes, which are enriched in heat-response and ribosomal protein genes, suggesting that they could be implicated in dynamic coordination of ribosome biogenesis. Chromatin immunoprecipitation experiments show that Corto and RPL12 bind hsp70 and are similarly recruited on gene body after heat shock. Hence, Corto and RPL12 could be involved together in regulation of gene transcription. We discuss whether pseudo-ribosomal complexes composed of various ribosomal proteins might participate in regulation of gene expression in connection with chromatin regulators.',
'date' => '2012-10-11',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/23071455',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '930',
'name' => 'The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts',
'authors' => 'Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J',
'description' => 'Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain and a Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts (MEFs) stably over-expressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the non-coding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2 and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches, showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression and the metabolic control of fibroblasts. ',
'date' => '2012-07-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22825849',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '352',
'name' => 'Mouse IgG SDS US en',
'language' => 'en',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-US-en-GHS_1_0.pdf',
'countries' => 'US',
'modified' => '2020-06-09 15:24:43',
'created' => '2020-06-09 15:24:43',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '350',
'name' => 'Mouse IgG SDS GB en',
'language' => 'en',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-GB-en-GHS_1_0.pdf',
'countries' => 'GB',
'modified' => '2020-06-09 15:23:14',
'created' => '2020-06-09 15:23:14',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '348',
'name' => 'Mouse IgG SDS ES es',
'language' => 'es',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-ES-es-GHS_1_0.pdf',
'countries' => 'ES',
'modified' => '2020-06-09 15:22:03',
'created' => '2020-06-09 15:22:03',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '347',
'name' => 'Mouse IgG SDS DE de',
'language' => 'de',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-DE-de-GHS_1_0.pdf',
'countries' => 'DE',
'modified' => '2020-06-09 15:21:25',
'created' => '2020-06-09 15:21:25',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '351',
'name' => 'Mouse IgG SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-JP-ja-GHS_1_0.pdf',
'countries' => 'JP',
'modified' => '2020-06-09 15:23:45',
'created' => '2020-06-09 15:23:45',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '346',
'name' => 'Mouse IgG SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-BE-nl-GHS_1_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-09 15:20:49',
'created' => '2020-06-09 15:20:49',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '345',
'name' => 'Mouse IgG SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-BE-fr-GHS_1_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-09 15:19:47',
'created' => '2020-06-09 15:19:47',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '349',
'name' => 'Mouse IgG SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-FR-fr-GHS_1_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-09 15:22:38',
'created' => '2020-06-09 15:22:38',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/mouse-igg-15-ug-15-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
)
$pro = array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/bioruptor-pico-sonication-device"><img src="/img/product/shearing_technologies/bioruptor_pico.jpg" alt="Bioruptor pico next gen sequencing " class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">B01060010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Bioruptor® Pico sonication device</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '1862',
'product_id' => '1959',
'related_id' => '1787'
),
'Image' => array(
(int) 0 => array(
'id' => '133',
'name' => 'product/shearing_technologies/bioruptor_pico.jpg',
'alt' => 'Bioruptor pico next gen sequencing ',
'modified' => '2020-11-27 10:00:30',
'created' => '2015-06-10 17:28:55',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1772',
'name' => 'product/shearing_technologies/B010600010.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 15:41:46',
'created' => '2018-02-14 15:41:46',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1773',
'name' => 'product/shearing_technologies/B010600010-2.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 16:18:03',
'created' => '2018-02-14 15:42:00',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1774',
'name' => 'product/shearing_technologies/B01060010-bioruptor-pico-cooler.jpg',
'alt' => 'Pico + cooler',
'modified' => '2018-03-06 11:01:33',
'created' => '2018-03-06 11:01:33',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '2896',
'antibody_id' => '328',
'name' => 'Mouse IgG ',
'description' => '<p>The negative Ctrl IgG from mouse has been extensively validated in chromatin immunoprecipitation assays (ChIP). It contains a spectrum of the IgG subclasses present in serum of healthy animals. This IgG preparation is intended for use as a negative control in ChIP experiments (but also in MeDIP, IF and other experiments) for specific antibodies made in mouse. The negative Ctrl IgG from mouse should be used for ChIP in parallel with specific antibody at the same concentration as the specific antibody.</p>',
'label1' => 'Validation data',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '100 µg',
'catalog_number' => 'C15400001-100',
'old_catalog_number' => '',
'sf_code' => 'C15400001-D001-000526',
'type' => 'FRE',
'search_order' => '',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'mouse-igg-100-ug',
'meta_title' => 'Mouse IgG (C15400001) | Diagenode',
'meta_keywords' => '',
'meta_description' => 'Mouse IgG - for use as a negative control in ChIP, MeDIP, IF and other experiments performed with specific antibodies made in mouse. Sample size available.',
'modified' => '2021-03-15 16:58:03',
'created' => '2017-05-17 15:25:18',
'ProductsGroup' => array(
'id' => '236',
'product_id' => '2896',
'group_id' => '211'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
'id' => '4167',
'product_id' => '1959',
'application_id' => '43'
)
)
$slugs = array(
(int) 0 => 'chip-qpcr-antibodies'
)
$applications = array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'locale' => 'eng'
)
$description = ''
$name = 'ChIP-qPCR (ab)'
$document = array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '2068',
'product_id' => '1959',
'document_id' => '38'
)
)
$sds = array(
'id' => '349',
'name' => 'Mouse IgG SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/Mouse_IgG/SDS-C15400001-Mouse_IgG-FR-fr-GHS_1_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-09 15:22:38',
'created' => '2020-06-09 15:22:38',
'ProductsSafetySheet' => array(
'id' => '681',
'product_id' => '1959',
'safety_sheet_id' => '349'
)
)
$publication = array(
'id' => '930',
'name' => 'The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology and the metabolome of fibroblasts',
'authors' => 'Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J',
'description' => 'Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC-domain-containing histone demethy-lase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain and a Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts (MEFs) stably over-expressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the non-coding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2 and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches, showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression and the metabolic control of fibroblasts. ',
'date' => '2012-07-23',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/22825849',
'doi' => '',
'modified' => '2015-07-24 15:38:58',
'created' => '2015-07-24 15:38:58',
'ProductsPublication' => array(
'id' => '1075',
'product_id' => '1959',
'publication_id' => '930'
)
)
$externalLink = ' <a href="http://www.ncbi.nlm.nih.gov/pubmed/22825849" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
×