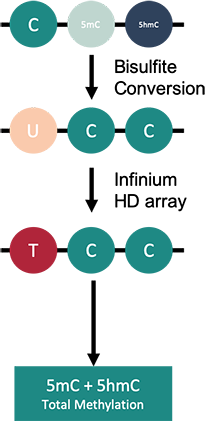
How to properly cite our product/service in your work We strongly recommend using this: Infinium MethylationEPIC Array Service (Hologic Diagenode Cat# G02090000). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Integrative network analysis of differentially methylated regions to study the impact of gestational weight gain on maternal metabolism and fetal-neonatal growth
Argentato P.P. et al.
Integrative network analysis (INA) is important for identifying gene modules or epigenetically regulated molecular pathways in diseases. This study evaluated the effect of excessive gestational weight gain (EGWG) on INA of differentially methylated regions, maternal metabolism and offspring growth. Brazilian women f... |
Making Biological Ageing Clocks Personal
Pusparum M. et al.
Background Age is the most important risk factor for the majority of human diseases. Addressing the impact of age-related diseases has become a priority in healthcare practice, leading to the exploration of innovative approaches, including the development of predictors to estimate biological age (so-called &l... |
Inhibition of Aurora kinase induces endogenous retroelements to induce a type I/III interferon response via RIG-I
Choy L. et al.
Type I interferon signaling is a crucial component of anti-viral immunity that has been linked to promoting the efficacy of some chemotherapeutic drugs. We developed a reporter system in HCT116 cells that detects activation of the endogenous IFI27 locus, an interferon (IFN) target gene. We screened a library of anno... |
Association of DNA methylation signatures with premature ageing andcardiovascular death in patients with end-stage kidney disease: a pilotepigenome-wide association study.
Sumida K. et al.
Patients with end-stage kidney disease (ESKD) display features of premature aging. There is strong evidence that changes in DNA methylation (DNAm) contribute to age-related pathologies; however, little is known about their association with premature aging and cardiovascular mortality in patients with ESKD. We assaye... |
Perturbed epigenetic transcriptional regulation in AML with IDHmutations causes increased susceptibility to NK cells.
Palau A. et al.
Isocitrate dehydrogenase (IDH) mutations are found in 20\% of acute myeloid leukemia (AML) patients. However, only 30-40\% of the patients respond to IDH inhibitors (IDHi). We aimed to identify a molecular vulnerability to tailor novel therapies for AML patients with IDH mutations. We characterized the transcription... |
In skeletal muscle and neural crest cells, SMCHD1 regulates biologicalpathways relevant for Bosma syndrome and facioscapulohumeral dystrophyphenotype.
Laberthonnière C. et al.
Many genetic syndromes are linked to mutations in genes encoding factors that guide chromatin organization. Among them, several distinct rare genetic diseases are linked to mutations in SMCHD1 that encodes the structural maintenance of chromosomes flexible hinge domain containing 1 chromatin-associated factor. In hu... |
Altered DNA methylation and gene expression predict disease severity inpatients with Aicardi-Goutières syndrome.
Garau J. et al.
Aicardi-Goutières Syndrome (AGS) is a rare neuro-inflammatory disease characterized by increased expression of interferon-stimulated genes (ISGs). Disease-causing mutations are present in genes associated with innate antiviral responses. Disease presentation and severity vary, even between patients with ident... |
DNA methylation aberrancy is a reliable prognostic tool in uveal melanoma
Soltysova A. et al.
Despite outstanding advances in understanding the genetic background of uveal melanoma (UM) development and prognosis, the role of DNA methylation reprogramming remains elusive. This study aims to clarify the extent of DNA methylation deregulation in the context of gene expression changes and its utility as a reliab... |
Decitabine increases neoantigen and cancer testis antigen expression toenhance T cell-mediated toxicity against glioblastoma.
Ma Ruichong et al.
BACKGROUND: Glioblastoma (GBM) is the most common and malignant primary brain tumour in adults. Despite maximal treatment, median survival remains dismal at 14-24 months. Immunotherapies, such as checkpoint inhibition, have revolutionised management of some cancers but have little benefit for GBM patients. This is, ... |
Enhanced cell deconvolution of peripheral blood using DNA methylation for high-resolution immune profiling
Lucas A Salas, Ze Zhang, Devin C Koestler, Rondi A Butler, Helen M Hansen, Annette M Molinaro, John K Wiencke, Karl T Kelsey, Brock C Christensen
DNA methylation microarrays can be employed to interrogate cell-type composition in complex tissues. Here, we expand reference-based deconvolution of blood DNA methylation to include 12 leukocyte subtypes (neutrophils, eosinophils, basophils, monocytes, B cells, CD4+ and CD8+ naïve and memory cells, natural kil... |
Interplay between Histone and DNA Methylation Seen through Comparative Methylomes in Rare Mendelian Disorders
Guillaume Velasco, Damien Ulveling,Sophie Rondeau,Pauline Marzin,Motoko Unoki,Valérie Cormier-Daire, Claire Francastel
DNA methylation (DNAme) profiling is used to establish specific biomarkers to improve the diagnosis of patients with inherited neurodevelopmental disorders and to guide mutation screening. In the specific case of mendelian disorders of the epigenetic machinery, it also provides the basis to infer mechanistic aspects... |
Genome-wide DNA methylation and transcriptome integration reveal distinct sex differences in skeletal muscle
Shanie Landen, Macsue Jacques , Danielle Hiam , Javier Alvarez, Nicholas R Harvey, Larisa M. Haupt, Lyn R, Griffiths, Kevin J Ashton, Séverine Lamon, Sarah Voisin, Nir Eynon
Nearly all human complex traits and diseases exhibit some degree of sex differences, and epigenetics contributes to these differences as DNA methylation shows sex differences in various tissues. However, skeletal muscle epigenetic sex differences remain largely unexplored, yet skeletal muscle displays distinct sex d... |
ZNF718, HOXA4, and ZFP57 are differentially methylated inperiodontitis in comparison with periodontal health: Epigenome-wide DNAmethylation pilot study.
Hernández H.G. et al.
OBJECTIVE: To investigate the differences in the epigenomic patterns of DNA methylation in peripheral leukocytes between patients with periodontitis and gingivally healthy controls evaluating its functional meaning by functional enrichment analysis. BACKGROUND: The DNA methylation profiling of peripheral leukocytes ... |
From methylation to myelination: epigenomic and transcriptomic profilingof chronic inactive demyelinated multiple sclerosis lesions
Tiane A. et al.
Introduction In the progressive phase of multiple sclerosis (MS), the hampered differentiation capacity of oligodendrocyte precursor cells (OPCs) eventually results in remyelination failure. We have previously shown that DNA methylation of Id2/Id4 is highly involved in OPC differentiation and remyelination. In this ... |