Download the manual
Download the manual
D-Plex Small RNA DNBSEQ™ Kit is a tool designed for the study of the small non-coding transcriptome. The kit is using the D-Plex technology to generate double-stranded DNA libraries ready to be used for the DNA single-strand circularization step required for DNBSEQ sequencing on MGI sequencers.
The D-Plex technology utilizes the innovative capture and amplification by tailing and switching, a ligation-free method for RNA library preparation from ultra-low input amounts, down to 10 pg for small RNAs and 100 pg for total RNAs. This innovative solution enables diverse and novel transcripts detection, even from challenging clinical samples such as liquid biopsies.
D-Plex Small RNA DNBSEQ™ Kit offers a time saving protocol that can be completed within 5 hours and requires minimal hands-on time. The library preparation takes place in a single tube, increasing the efficiency tremendously. This ensures high technical reliability and reproducibility.
D-Plex Small RNA DNBSEQ™ Kit includes all buffers and enzymes necessary for the library preparation. Specific D-Plex DNBSEQ Barcodes were designed and validated to fit the D-Plex technology and are available separately:
D-Plex is also available for Illumina sequencing, check here!
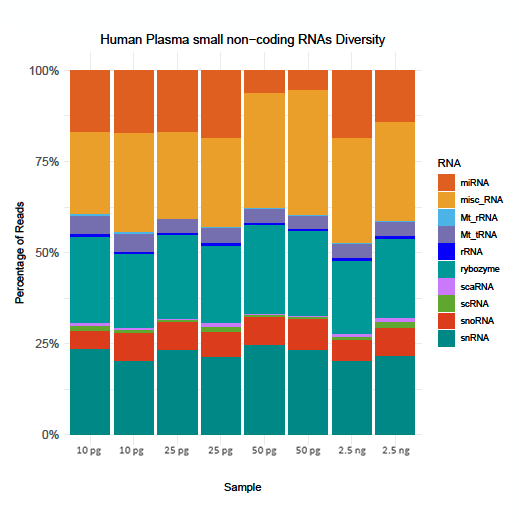
Diverse and novel transcript detection
The D-Plex Small RNA DNBSEQ protocol generates complex RNA libraries deciphering the wide diversity of small non-coding RNA spectrum (including miRNAs, snoRNAs, snRNAs) in human plasma samples.
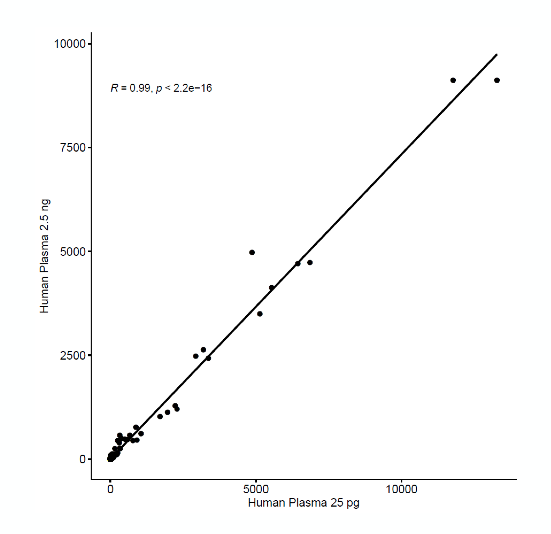
Ultra-low input performance
Sequencing data from circulating RNA samples of two input amounts (25 pg and 2.5 ng) were highly correlated (R = 0.99) when compared using Pearson correlation coefficient.
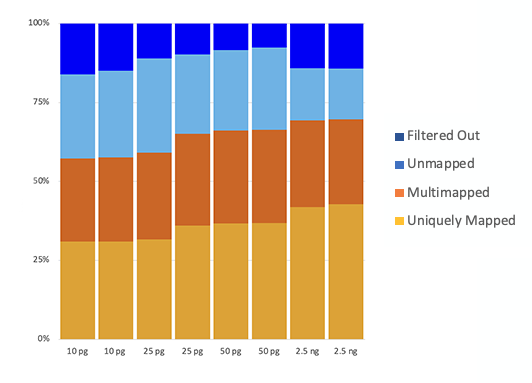
High mapping efficiency
The D-Plex Small RNA DNBSEQ kit is compatible with clinical-relevant samples, such as human plasma, and ultra low range of circulating RNA input (down to 10 pg) and exhibits good read mapping of sequencing reads (up to 70% mapping rate).
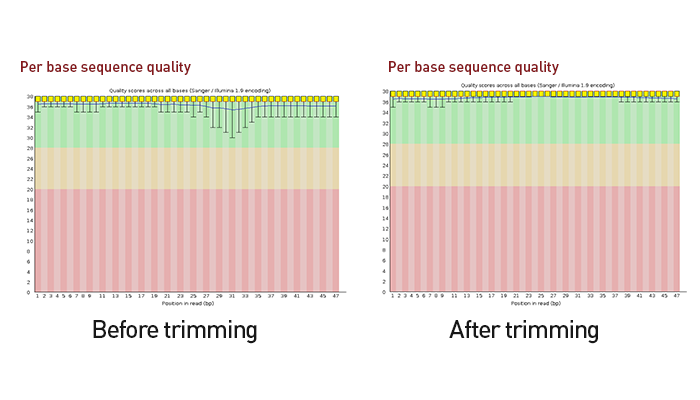
High quality DNBSEQ sequencing solution
The D-Plex Small RNA DNBSEQ kit combines our best-in-class RNA library preparation – D-Plex technology – with MGI’s high-quality, cost-effective, DNA nanoballs – DNBSEQ – sequencing solution, creating a unified platform to support high quality small RNA sequencing.