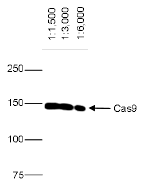
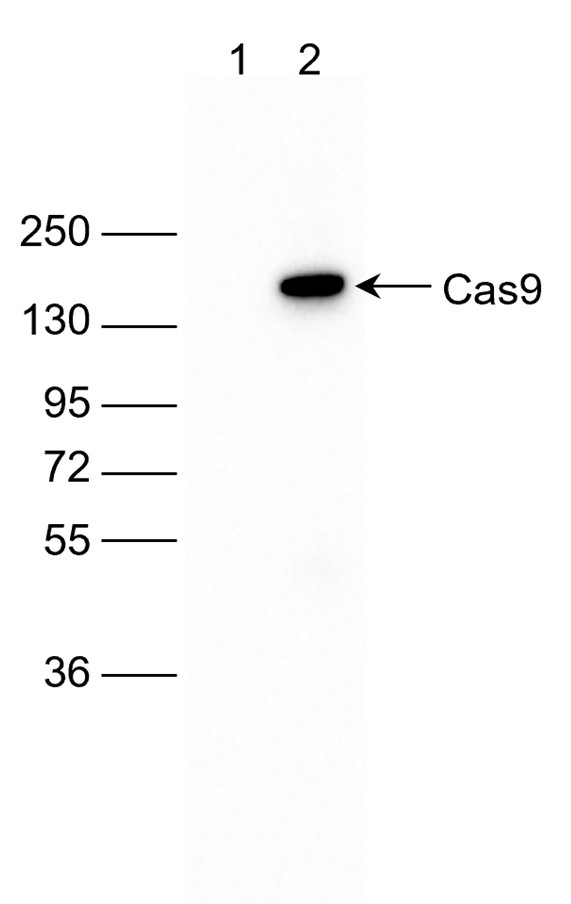
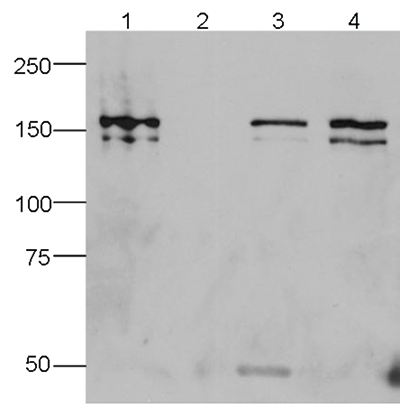
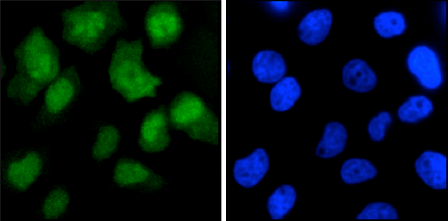
How to properly cite our product/service in your work We strongly recommend using this: CRISPR/Cas9 Antibody – clone 7A9 (Hologic Diagenode Cat# C15200203-50 Lot# 006). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Loss of tumor suppressors promotes inflammatory tumor microenvironment and enhances LAG3+T cell mediated immune suppression
Zahraeifard, S. et al.
Low response rate, treatment relapse, and resistance remain key challenges for cancer treatment with immune checkpoint blockade (ICB). Here we report that loss of specific tumor suppressors (TS) induces an inflammatory response and promotes an immune suppressive tumor microenvironment. Importantly, low expression of... |
A kinesin-based approach for inducing chromosome-specific mis-segregationin human cells.
Truong M.A. et al.
Various cancer types exhibit characteristic and recurrent aneuploidy patterns. The origins of these cancer type-specific karyotypes are still unknown, partly because introducing or eliminating specific chromosomes in human cells still poses a challenge. Here, we describe a novel strategy to induce mis-segregation of... |
MICAL1 regulates actin cytoskeleton organization, directional cellmigration and the growth of human breast cancer cells as orthotopicxenograft tumours.
McGarry David J et al.
The Molecule Interacting with CasL 1 (MICAL1) monooxygenase has emerged as an important regulator of cytoskeleton organization via actin oxidation. Although filamentous actin (F-actin) increases MICAL1 monooxygenase activity, hydrogen peroxide (HO) is also generated in the absence of F-actin, suggesting that diffusi... |
CRISPR-based gene knockout screens reveal deubiquitinases involved in HIV-1 latency in two Jurkat cell models.
Rathore A, Iketani S, Wang P, Jia M, Sahi V, Ho DD
The major barrier to a HIV-1 cure is the persistence of latent genomes despite treatment with antiretrovirals. To investigate host factors which promote HIV-1 latency, we conducted a genome-wide functional knockout screen using CRISPR-Cas9 in a HIV-1 latency cell line model. This screen identified IWS1, POLE3, POLR1... |
DAPK1 loss triggers tumor invasion in colorectal tumor cells.
Steinmann S, Kunze P, Hampel C, Eckstein M, Bertram Bramsen J, Muenzner JK, Carlé B, Ndreshkjana B, Kemenes S, Gasparini P, Friedrich O, Andersen C, Geppert C, Wang S, Eyupoglu I, Bäuerle T, Hartmann A, Schneider-Stock R
Colorectal cancer (CRC) is one of the leading cancer-related causes of death worldwide. Despite the improvement of surgical and chemotherapeutic treatments, as of yet, the disease has not been overcome due to metastasis to distant organs. Hence, it is of great relevance to understand the mechanisms responsible for m... |
Optimization of CRISPR/Cas9 Delivery to Human Hematopoietic Stem and Progenitor Cells for Therapeutic Genomic Rearrangements.
Lattanzi A, Meneghini V, Pavani G, Amor F, Ramadier S, Felix T, Antoniani C, Masson C, Alibeu O, Lee C, Porteus MH, Bao G, Amendola M, Mavilio F, Miccio A
Editing the β-globin locus in hematopoietic stem cells is an alternative therapeutic approach for gene therapy of β-thalassemia and sickle cell disease. Using the CRISPR/Cas9 system, we genetically modified human hematopoietic stem and progenitor cells (HSPCs) to mimic the large rearrangements in the &beta... |
Can Mitochondrial DNA be CRISPRized: Pro and Contra.
Loutre R, Heckel AM, Smirnova A, Entelis N, Tarassov I
Mitochondria represent a chimera of macromolecules encoded either in the organellar genome, mtDNA, or in the nuclear one. If the pathway of protein targeting to different sub-compartments of mitochondria was relatively well studied, import of small noncoding RNAs into mammalian mitochondria still awaits mechanistic ... |
In trans paired nicking triggers seamless genome editing without double-stranded DNA cutting
Chen X. et al.
Precise genome editing involves homologous recombination between donor DNA and chromosomal sequences subjected to double-stranded DNA breaks made by programmable nucleases. Ideally, genome editing should be efficient, specific, and accurate. However, besides constituting potential translocation-initiating lesions, d... |
Highly efficient gene inactivation by adenoviral CRISPR/Cas9 in human primary cells
Voets O. et al.
Phenotypic assays using human primary cells are highly valuable tools for target discovery and validation in drug discovery. Expression knockdown (KD) of such targets in these assays allows the investigation of their role in models of disease processes. Therefore, efficient and fast modes of protein KD in phenotypic... |
Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer
Bailey SD et al.
Sustained expression of the estrogen receptor-α (ESR1) drives two-thirds of breast cancer and defines the ESR1-positive subtype. ESR1 engages enhancers upon estrogen stimulation to establish an oncogenic expression program1. Somatic copy number alterations involving the ESR1 gene occur in approximately 1% of E... |
The Development of a Viral Mediated CRISPR/Cas9 System with Doxycycline Dependent gRNA Expression for Inducible In vitro and In vivo Genome Editing.
de Solis CA et al.
The RNA-guided Cas9 nuclease, from the type II prokaryotic Clustered Regularly Interspersed Short Palindromic Repeats (CRISPR) adaptive immune system, has been adapted and utilized by scientists to edit the genomes of eukaryotic cells. Here, we report the development of a viral mediated CRISPR/Cas9 system that can b... |
CRISPR-Mediated Gene Targeting of Human Induced Pluripotent Stem Cells
Susan M. Byrne, George M. Church
CRISPR/Cas9 nuclease systems can create double-stranded DNA breaks at specific sequences to efficiently and precisely disrupt, excise, mutate, insert, or replace genes. However, human embryonic stem cells and induced pluripotent stem cells (iPSCs) are more difficult to transfect and less resilient to DNA damage than... |
A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing
Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, Liu Y
CRISPR/Cas has emerged as potent genome editing technology and has successfully been applied in many organisms, including several plant species. However, delivery of genome editing reagents remains a challenge in plants. Here, we report a virus-based guide RNA (gRNA) delivery system for CRISPR/Cas9 mediated plant ge... |
Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection.
Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD
CRISPR-Cas9 systems provide a platform for high efficiency genome editing that are enabling innovative applications of mammalian cell engineering. However, the delivery of Cas9 and synthesis of guide RNA (gRNA) remain as steps that can limit overall efficiency and ease of use. Here we describe methods for rapid synt... |
A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage.
van Sluis M, McStay B
DNA double-strand breaks (DSBs) are repaired by two main pathways: nonhomologous end-joining and homologous recombination (HR). Repair pathway choice is thought to be determined by cell cycle timing and chromatin context. Nucleoli, prominent nuclear subdomains and sites of ribosome biogenesis, form around nucleolar ... |
Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis.
Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H, Zhang F, Sharp PA
Genetic screens are powerful tools for identifying genes responsible for diverse phenotypes. Here we describe a genome-wide CRISPR/Cas9-mediated loss-of-function screen in tumor growth and metastasis. We mutagenized a non-metastatic mouse cancer cell line using a genome-scale library with 67,405 single-guide RNAs (s... |
Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells.
Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang CJ, Esteban CR, Young J, Izpisua Belmonte JC
To combat hostile viruses, bacteria and archaea have evolved a unique antiviral defense system composed of clustered regularly interspaced short palindromic repeats (CRISPRs), together with CRISPR-associated genes (Cas). The CRISPR/Cas9 system develops an adaptive immune resistance to foreign plasmids and viruses by... |
An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo.
Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O'Connor L, Milla L, Wilcox S, Tai L, Strasser A, Herold MJ
The CRISPR/Cas9 technology enables the introduction of genomic alterations into almost any organism; however, systems for efficient and inducible gene modification have been lacking, especially for deletion of essential genes. Here, we describe a drug-inducible small guide RNA (sgRNA) vector system allowing for ubiq... |
An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo
Aubrey BJ et al.
The CRISPR/Cas9 technology enables the introduction of genomic alterations into almost any organism; however, systems for efficient and inducible gene modification have been lacking, especially for deletion of essential genes. Here, we describe a drug-inducible small guide RNA (sgRNA) vector system allowing for ubiq... |