- Highly optimized protocol for chromatin preparation prior to ChIP on transcription factors and non-histone proteins
- SDS concentration optimized for the workflow for TFs
- Validated for cells, tissue and FFPE samples
- Preserves the epitopes
- Validated with the Bioruptor ultrasonicator
- Quality of chromatin sample confirmed by ChIP-seq
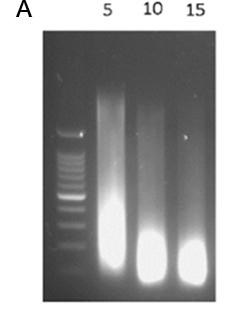
Figure 1. Optimal chromatin shearing profile
HeLa cells were fixed with formaldehyde for 10 min and chromatin was prepared according to Diagenode’s Chromatin EasyShear Kit - Low SDS (Cat. No. C01020013). Samples were sonicated for 5-10-15 cycles of 30” ON/30” OFF as indicated with Bioruptor Pico using 1.5 ml Bioruptor microtubes with caps (Cat. No. C30010016) followed by de-crosslinking and DNA purification. The fragment size was assessed using agarose gel electrophoresis. A 100 bp ladder was loaded as the size standard.


Figure 2. Chromatin precipitation
Sheared chromatin (obtained with the Chromatin EasyShear Kit – Low SDS) has been used for immunoprecipitation with CTCF and IgG (negative control) antibodies. Quantitative PCR was performed with positive (H19) and negative (Myoglobine exon 2) control regions. The Figure 2 shows the recovery expressed as % of input (the relative amount of immunoprecipitated DNA compared to input DNA (panel A) and as enrichment fold of positive locus over negative (panel B).
