CATSは、核酸の3 '末端を捕捉するポリヌクレオチドテーリングおよびcDNA合成のための5'末端を捕捉するMMLV-RTのテンプレートスイッチング能力に依存します。
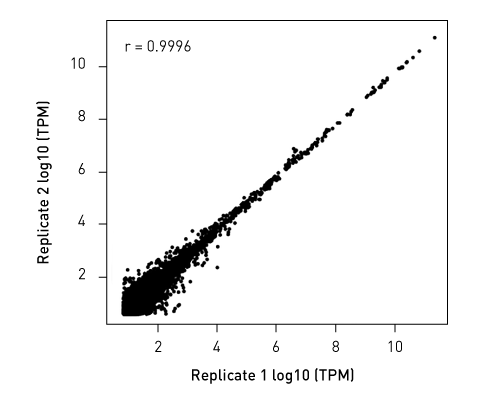
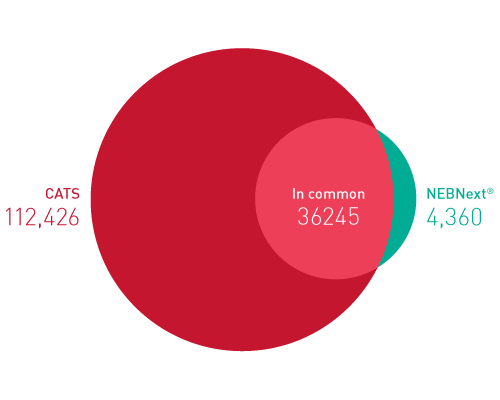
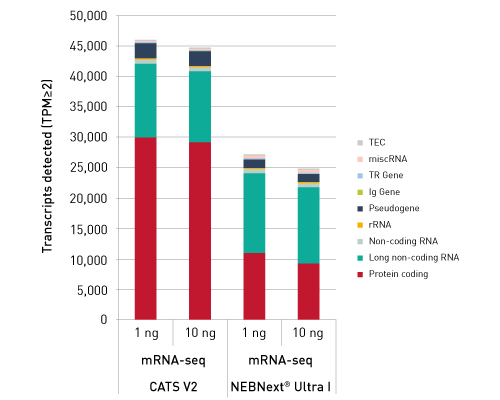
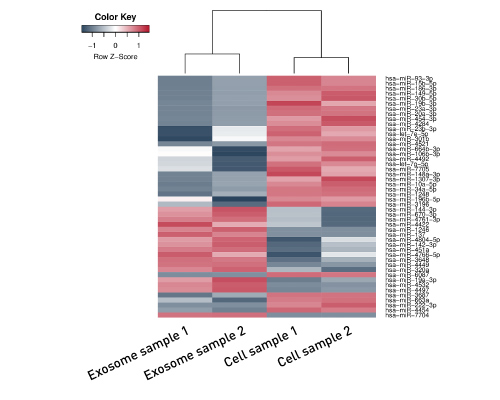
Diagenodeキットはわずか数時間で10pgという低いRNA入力からIlluminaに互換のready-to-sequenceライブラリーを生成します。 ライブラリーは、起点の鎖特異性を保持します。他社製品とは異なり、当社のCATS RNA-seqキットはライゲーションに基づく方法と比較してより高いライブラリー効率を示します。 CATS法は、より多くの非コードRNAの正確な同定を可能にするだけでなく、より良好な鋳型捕捉からより高い複雑性、低い増幅要件から最小バイアスを提供します。