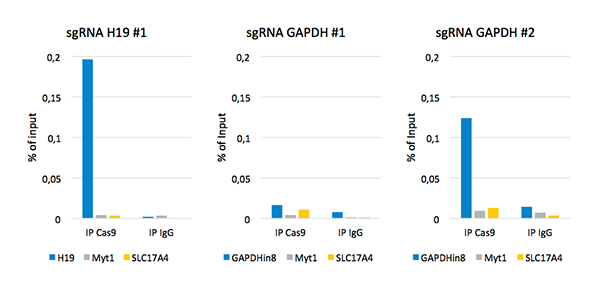
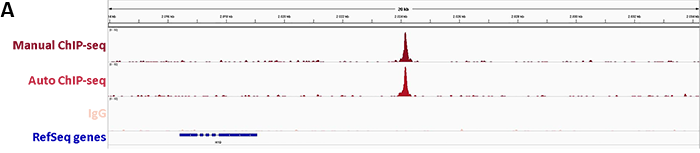
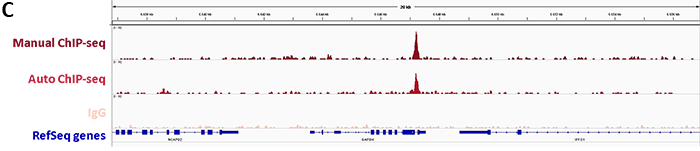
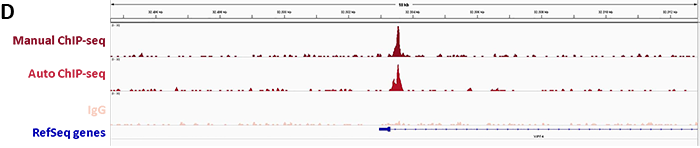
ChIP was performed on sheared chromatin from 4,000,000 HEK293T cells using the iDeal ChIP-seq Kit for Transcription Factors, 5 µl of the polyclonal Cas9 antibody and 1 µg of the negative control IgG. Primers specific for the human H19, GAPDH, Myt1 and SLC17A4 were used for the qPCR. The figure shows the recovery, expressed as a percent of input (the relative amount of immunoprecipitated DNA compared to input after qPCR analysis).