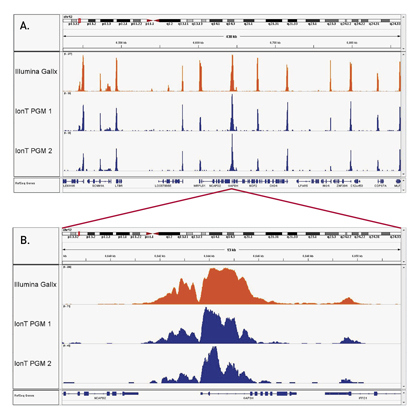
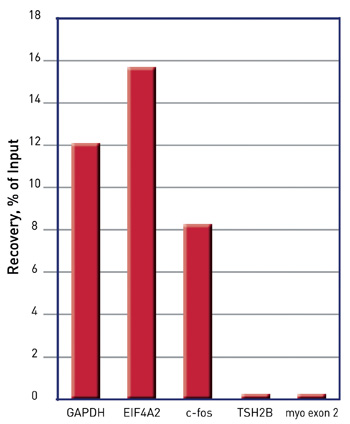
How to properly cite our product/service in your work We strongly recommend using this: Auto iDeal ChIP-seq kit for Histones (Hologic Diagenode Cat# C01010171). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
K27M in canonical and noncanonical H3 variants occurs in distinctoligodendroglial cell lineages in brain midline gliomas.
Jessa Selin et al.
Canonical (H3.1/H3.2) and noncanonical (H3.3) histone 3 K27M-mutant gliomas have unique spatiotemporal distributions, partner alterations and molecular profiles. The contribution of the cell of origin to these differences has been challenging to uncouple from the oncogenic reprogramming induced by the mutation. Here... |
Effects of GSK-J4 on JMJD3 Histone Demethylase in Mouse Prostate Cancer Xenografts
Sanchez A. et al.
Background/aim: Histone methylation status is required to control gene expression. H3K27me3 is an epigenetic tri-methylation modification to histone H3 controlled by the demethylase JMJD3. JMJD3 is dysregulated in a wide range of cancers and has been shown to control the expression of a specific growth-modulato... |
Sp1-Induced SETDB1 Overexpression Transcriptionally InhibitsHPGD in a β-Catenin-Dependent Manner and Promotes theProliferation and Metastasis of Gastric Cancer
Fan Y. et al.
Gastric cancer (GC) has high morbidity and mortality, the cure rate of surgical treatment and drug chemotherapy is not ideal. Therefore, development of new treatment strategies is necessary. We aimed to identify the mechanism underlying Sp1 regulation of GC progression. |
Epigenetic Drifts during Long-Term Intestinal OrganoidCulture
Thalheim T. et al.
Organoids retain the morphological and molecular patterns of their tissue of origin, are self-organizing, relatively simple to handle and accessible to genetic engineering. Thus, they represent an optimal tool for studying the mechanisms of tissue maintenance and aging. Long-term expansion under standard growth cond... |
Chromatin dysregulation associated with NSD1 mutation in head and necksquamous cell carcinoma.
Farhangdoost, Nargess et al.
Chromatin dysregulation has emerged as an important mechanism of oncogenesis. To develop targeted treatments, it is important to understand the transcriptomic consequences of mutations in chromatin modifier genes. Recently, mutations in the histone methyltransferase gene nuclear receptor binding SET domain protein 1... |
Postoperative abdominal sepsis induces selective and persistent changes inCTCF binding within the MHC-II region of human monocytes.
Siegler B. et al.
BACKGROUND: Postoperative abdominal infections belong to the most common triggers of sepsis and septic shock in intensive care units worldwide. While monocytes play a central role in mediating the initial host response to infections, sepsis-induced immune dysregulation is characterized by a defective antigen present... |
Histone H3.3G34-Mutant Interneuron Progenitors Co-opt PDGFRA for Gliomagenesis.
Chen C. et al.
Histone H3.3 glycine 34 to arginine/valine (G34R/V) mutations drive deadly gliomas and show exquisite regional and temporal specificity, suggesting a developmental context permissive to their effects. Here we show that 50\% of G34R/V tumors (n = 95) bear activating PDGFRA mutations that display strong selection... |
EZH2 and KDM6B Expressions Are Associated with Specific EpigeneticSignatures during EMT in Non Small Cell Lung Carcinomas.
Lachat C. et al.
The role of Epigenetics in Epithelial Mesenchymal Transition (EMT) has recently emerged. Two epigenetic enzymes with paradoxical roles have previously been associated to EMT, EZH2 (Enhancer of Zeste 2 Polycomb Repressive Complex 2 (PRC2) Subunit), a lysine methyltranserase able to add the H3K27me3 mark, and the hist... |
Digging Deeper into Breast Cancer Epigenetics: Insights from ChemicalInhibition of Histone Acetyltransferase TIP60 .
Idrissou, Mouhamed and Lebert, Andre and Boisnier, Tiphanie and Sanchez,Anna and Houfaf Khoufaf, Fatma Zohra and Penault-Llorca, Frédérique andBignon, Yves-Jean and Bernard-Gallon, Dominique
Breast cancer is often sporadic due to several factors. Among them, the deregulation of epigenetic proteins may be involved. TIP60 or KAT5 is an acetyltransferase that regulates gene transcription through the chromatin structure. This pleiotropic protein acts in several cellular pathways by acetylating proteins. RNA... |
Human papillomavirus type 38 alters wild-type p53 activity to promote cellproliferation via the downregulation of integrin alpha 1 expression.
Romero-Medina, MC and Venuti, A and Melita, G and Robitaille, A andCeraolo, MG and Pacini, L and Sirand, C and Viarisio, D and Taverniti, Vand Gupta, P and Scalise, M and Indiveri, C and Accardi, R and Tommasino, M
Tumor suppressors can exert pro-proliferation functions in specific contexts. In the beta human papillomavirus type 38 (HPV38) experimental model, the viral proteins E6 and E7 promote accumulation of a wild-type (WT) p53 form in human keratinocytes (HKs), promoting cellular proliferation. Inactivation of p53 by diff... |
The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape.
Weinberg DN, Papillon-Cavanagh S, Chen H, Yue Y, Chen X, Rajagopalan KN, Horth C, McGuire JT, Xu X, Nikbakht H, Lemiesz AE, Marchione DM, Marunde MR, Meiners MJ, Cheek MA, Keogh MC, Bareke E, Djedid A, Harutyunyan AS, Jabado N, Garcia BA, Li H, Allis CD,
Enzymes that catalyse CpG methylation in DNA, including the DNA methyltransferases 1 (DNMT1), 3A (DNMT3A) and 3B (DNMT3B), are indispensable for mammalian tissue development and homeostasis. They are also implicated in human developmental disorders and cancers, supporting the critical role of DNA methylation in the ... |
Pervasive H3K27 Acetylation Leads to ERV Expression and a Therapeutic Vulnerability in H3K27M Gliomas.
Krug B, De Jay N, Harutyunyan AS, Deshmukh S, Marchione DM, Guilhamon P, Bertrand KC, Mikael LG, McConechy MK, Chen CCL, Khazaei S, Koncar RF, Agnihotri S, Faury D, Ellezam B, Weil AG, Ursini-Siegel J, De Carvalho DD, Dirks PB, Lewis PW, Salomoni P, Lupie
High-grade gliomas defined by histone 3 K27M driver mutations exhibit global loss of H3K27 trimethylation and reciprocal gain of H3K27 acetylation, respectively shaping repressive and active chromatin landscapes. We generated tumor-derived isogenic models bearing this mutation and show that it leads to pervasive H3K... |
H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis.
Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, Deshmukh S, Chen CCL, Belle J, Mikael LG, Marchione DM, Li R, Nikbakht H, Hu B, Cagnone G, Cheung WA, Mohammadnia A, Bechet D, Faury D, McConechy MK, Pathania M, Jain SU, Ellezam B,
Lys-27-Met mutations in histone 3 genes (H3K27M) characterize a subgroup of deadly gliomas and decrease genome-wide H3K27 trimethylation. Here we use primary H3K27M tumor lines and isogenic CRISPR-edited controls to assess H3K27M effects in vitro and in vivo. We find that whereas H3K27me3 and H3K27me2 are normally d... |
PRC2 targeting is a therapeutic strategy for EZ score defined high-risk multiple myeloma patients and overcome resistance to IMiDs.
Herviou L, Kassambara A, Boireau S, Robert N, Requirand G, Müller-Tidow C, Vincent L, Seckinger A, Goldschmidt H, Cartron G, Hose D, Cavalli G, Moreaux J
BACKGROUND: Multiple myeloma (MM) is a malignant plasma cell disease with a poor survival, characterized by the accumulation of myeloma cells (MMCs) within the bone marrow. Epigenetic modifications in MM are associated not only with cancer development and progression, but also with drug resistance. METHODS: We ident... |
SIRT1-dependent epigenetic regulation of H3 and H4 histone acetylation in human breast cancer
Khaldoun Rifaï et al.
Breast cancer is the most frequently diagnosed malignancy in women worldwide. It is well established that the complexity of carcinogenesis involves profound epigenetic deregulations that contribute to the tumorigenesis process. Deregulated H3 and H4 acetylated histone marks are amongst those alterations. Sirtuin-1 (... |
TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors
Ancey P.B. et al.
Understanding the processes that govern liver progenitor cell differentiation has important implications for the design of strategies targeting chronic liver diseases, whereby regeneration of liver tissue is critical. Although DNA methylation (5mC) and hydroxymethylation (5hmC) are highly dynamic during early embryo... |
Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells
Chen L. et al.
Characterizing the multifaceted contribution of genetic and epigenetic factors to disease phenotypes is a major challenge in human genetics and medicine. We carried out high-resolution genetic, epigenetic, and transcriptomic profiling in three major human immune cell types (CD14+ monocytes, CD16+ neutrophils, ... |
H4K5 histone acetylation of BRG1 is associated with heroin administration rather than addiction
Xu L et al.
Diacetylmorphine hydrochloride (heroin) addiction is a chronic relapsing brain disorder that is a heavy public health burden worldwide. Brm/SWI2-related gene-1 (BRG1) is a tumor suppressor gene that can influence embryogenesis and the development of the cerebellum. The current study aimed to investigate the effect o... |