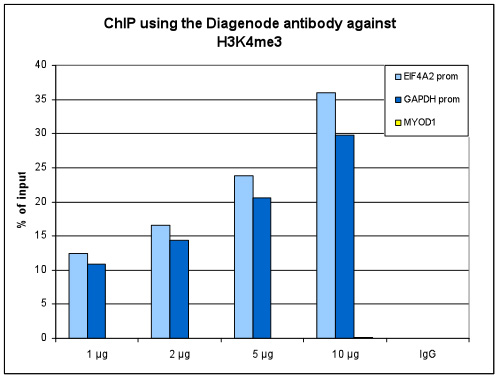
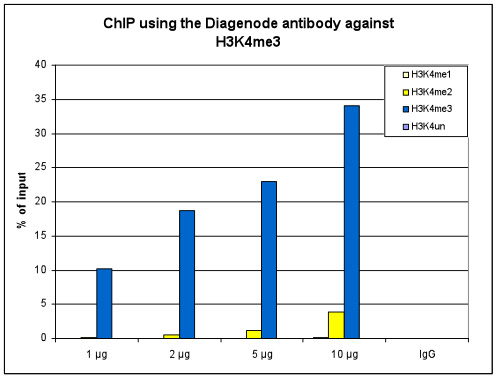
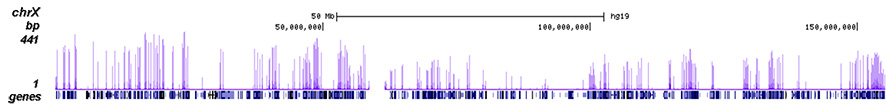
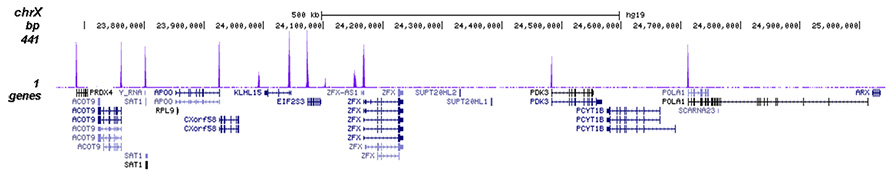
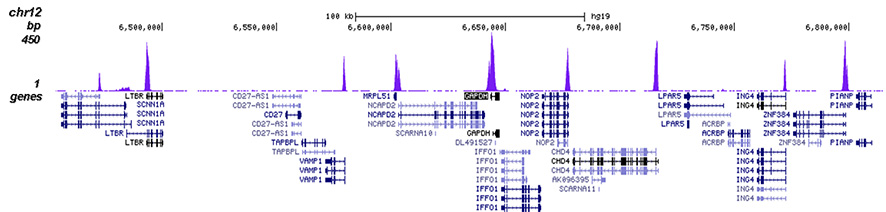
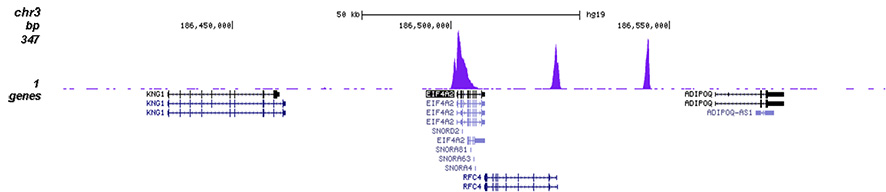
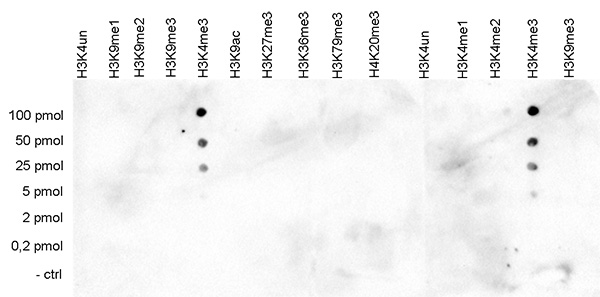
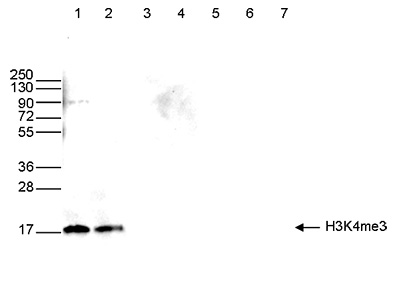
How to properly cite our product/service in your work We strongly recommend using this: H3K4me3 polyclonal antibody (Hologic Diagenode Cat# C15410030 Lot# 002). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
SUMO protease FUG1, histone reader AL3 and the PRC1 Complex areintegral to repeat-expansion induced epigenetic silencing in Arabidopsisthaliana
Sureshkumar S. et al.
Epigenetic gene silencing induced by expanded repeats can cause diverse phenotypes ranging from severe growth defects in plants to genetic diseases such as Friedreich’s ataxia in humans1. The molecular mechanisms underlying repeat expansion-induced epigenetic silencing remain largely unknown2,3. Using a plant ... |
Temporal modification of H3K9/14ac and H3K4me3 histone marksmediates mechano-responsive gene expression during the accommodationprocess in poplar
Ghosh R. et al.
Plants can attenuate their molecular response to repetitive mechanical stimulation as a function of their mechanical history. For instance, a single bending of stem is sufficient to attenuate the gene expression in poplar plants to the subsequent mechanical stimulation, and the state of desensitization can last for ... |
DNA sequence and chromatin modifiers cooperate to confer epigeneticbistability at imprinting control regions.
Butz S. et al.
Genomic imprinting is regulated by parental-specific DNA methylation of imprinting control regions (ICRs). Despite an identical DNA sequence, ICRs can exist in two distinct epigenetic states that are memorized throughout unlimited cell divisions and reset during germline formation. Here, we systematically study the ... |
Altered Chromatin States Drive Cryptic Transcription in AgingMammalian Stem Cells.
McCauley Brenna S et al.
A repressive chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation suppresses cryptic transcription in embryonic stem cells. Cryptic transcription is elevated with age in yeast and nematodes, and reducing it extends yeast lifespan, though whether this occurs in mammals is unk... |
Age-associated cryptic transcription in mammalian stem cells is linked topermissive chromatin at cryptic promoters
McCauley B. S. et al.
Suppressing spurious cryptic transcription by a repressive intragenic chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation is critical for maintaining self-renewal capacity in mouse embryonic stem cells. In yeast and nematodes, such cryptic transcription is elevated with age... |
Identification of Novel Molecular Markers of Human Th17 Cells.
Sałkowska A, Karaś K, Karwaciak I, Walczak-Drzewiecka A, Krawczyk M, Sobalska-Kwapis M, Dastych J, Ratajewski M
Th17 cells are important players in host defense against pathogens such as , , and . Th17 cell-mediated inflammation, under certain conditions in which balance in the immune system is disrupted, is the underlying pathogenic mechanism of certain autoimmune disorders, e.g., rheumatoid arthritis, Graves' disease, multi... |
The Chromatin Factor HNI9 and ELONGATED HYPOCOTYL5 Maintain ROS Homeostasis under High Nitrogen Provision.
Bellegarde F, Maghiaoui A, Boucherez J, Krouk G, Lejay L, Bach L, Gojon A, Martin A
Reactive oxygen species (ROS) can accumulate in cells at excessive levels, leading to unbalanced redox states and to potential oxidative stress, which can have damaging effects on the molecular components of plant cells. Several environmental conditions have been described as causing an elevation of ROS production i... |
Polycomb Repressive Complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1.
Bellegarde F, Herbert L, Séré D, Caillieux E, Boucherez J, Fizames C, Roudier F, Gojon A, Martin A
PRC2 is a major regulator of gene expression in eukaryotes. It catalyzes the repressive chromatin mark H3K27me3, which leads to very low expression of target genes. NRT2.1, which encodes a key root nitrate transporter in Arabidopsis, is targeted by H3K27me3, but the function of PRC2 on NRT2.1 remains unclear. Here, ... |
Interplay of cell–cell contacts and RhoA/MRTF‐A signaling regulates cardiomyocyte identity
Dorn et al
Cell–cell and cell–matrix interactions guide organ development and homeostasis by controlling lineage specification and maintenance, but the underlying molecular principles are largely unknown. Here, we show that in human developing cardiomyocytes cell–cell contacts at the intercalated disk connect... |
Neonatal exposure to hyperoxia leads to persistent disturbances in pulmonary histone signatures associated with NOS3 and STAT3 in a mouse model.
Chao CM, van den Bruck R, Lork S, Merkle J, Krampen L, Weil PP, Aydin M, Bellusci S, Jenke AC, Postberg J
Background: Early pulmonary oxygen exposure is one of the most important factors implicated in the development of bronchopulmonary dysplasia (BPD). Methods: Here, we analyzed short- and long-term effects of neonatal hyperoxia on NOS3 and STAT3 expression and corresponding epigenetic signatures using a hyperoxia-base... |
Rapid Communication: The correlation between histone modifications and expression of key genes involved in accumulation of adipose tissue in the pig.
Kociucka B. et al.
Histone modification is a well-known epigenetic mechanism involved in regulation of gene expression; however, it has been poorly studied in adipose tissues of the pig. Understanding the molecular background of adipose tissue development and function is essential for improving production efficiency and meat quality. ... |
Lhx2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11
Muralidharan B. et al.
n the developing cerebral cortex, sequential transcriptional programs take neuroepithelial cells from proliferating progenitors to differentiated neurons with unique molecular identities. The regulatory changes that occur in the chromatin of the progenitors are not well understood. During deep layer neurogenesis, we... |
Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape
Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G
BACKGROUND:
Previously, a role was demonstrated for transcription in the acquisition of DNA methylation at imprinted control regions in oocytes. Definition of the oocyte DNA methylome by whole genome approaches revealed that the majority of methylated CpG islands are intragenic and gene bodies are hypermethylated... |
CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.
Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC
BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the unde... |
Paclitaxel resistance increases oncolytic adenovirus efficacy via upregulated CAR expression and dysfunctional cell cycle control.
Ingemarsdotter CK, Tookman LA, Browne A, Pirlo K, Cutts R, Chelela C, Khurrum KF, Leung EY, Dowson S, Webber L, Khan I, Ennis D, Syed N, Crook TR, Brenton JD, Lockley M, McNeish IA
Resistance to paclitaxel chemotherapy frequently develops in ovarian cancer. Oncolytic adenoviruses are a novel therapy for human malignancies that are being evaluated in early phase trials. However, there are no reliable predictive biomarkers for oncolytic adenovirus activity in ovarian cancer. We investigated the ... |
p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6.
Phalke S, Mzoughi S, Bezzi M, Jennifer N, Mok WC, Low DH, Thike AA, Kuznetsov VA, Tan PH, Voorhoeve PM, Guccione E
p21 is a potent cyclin-dependent kinase inhibitor that plays a role in promoting G1 cell cycle arrest and cellular senescence. Consistent with this role, p21 is a downstream target of several tumour suppressors and oncogenes, and it is downregulated in the majority of tumours, including breast cancer. Here, we repor... |
Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.
Henckel A, Chebli K, Kota SK, Arnaud P, Feil R
Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation ... |