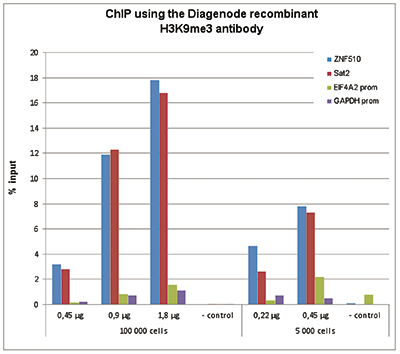
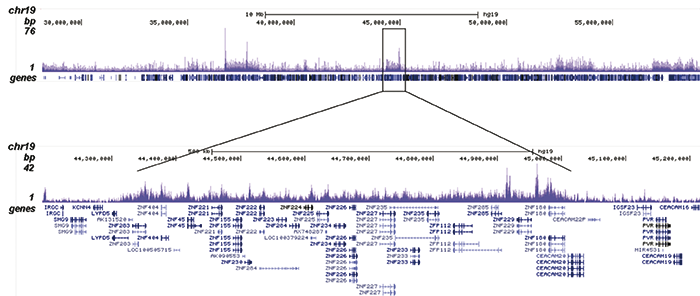
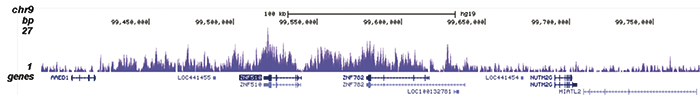
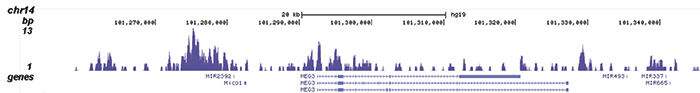
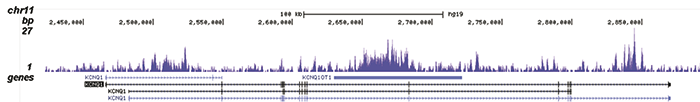
How to properly cite our product/service in your work We strongly recommend using this: H3K9me3 Antibody - ChIP-seq Grade (sample size) (Hologic Diagenode Cat# C15500003-10 Lot# 001). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
H3K9 dimethylation safeguards cancer cells against activation of theinterferon pathway.
Hansen A. M. et al.
Activation of interferon genes constitutes an important anticancer pathway able to restrict proliferation of cancer cells. Here, we demonstrate that the H3K9me3 histone methyltransferase (HMT) suppressor of variegation 3-9 homolog 1 (SUV39H1) is required for the proliferation of acute myeloid leukemia (AML) and find... |
SUV39 SET domains mediate crosstalk of heterochromatic histone marks.
Stirpe, Alessandro and Guidotti, Nora and Northall, Sarah Jand Kilic, Sinan and Hainard, Alexandre and Vadas, Oscar andFierz, Beat and Schalch, Thomas
The SUV39 class of methyltransferase enzymes deposits histone H3 lysine 9 di- and trimethylation (H3K9me2/3), the hallmark of constitutive heterochromatin. How these enzymes are regulated to mark specific genomic regions as heterochromatic is poorly understood. Clr4 is the sole H3K9me2/3 methyltransferase in the fis... |
A conserved RNA degradation complex required for spreading and epigenetic inheritance of heterochromatin.
Shipkovenska G, Durango A, Kalocsay M, Gygi SP, Moazed D
Heterochromatic domains containing histone H3 lysine 9 methylation (H3K9me) can be epigenetically inherited independently of underlying DNA sequence. To gain insight into the mechanisms that mediate epigenetic inheritance, we used a inducible heterochromatin formation system to perform a genetic screen for mutations... |
Genomic Profiling by ALaP-Seq Reveals Transcriptional Regulation by PML Bodies through DNMT3A Exclusion.
Kurihara M, Kato K, Sanbo C, Shigenobu S, Ohkawa Y, Fuchigami T, Miyanari Y
The promyelocytic leukemia (PML) body is a phase-separated nuclear structure physically associated with chromatin, implying its crucial roles in genome functions. However, its role in transcriptional regulation is largely unknown. We developed APEX-mediated chromatin labeling and purification (ALaP) to identify the ... |
Abo1 is required for the H3K9me2 to H3K9me3 transition in heterochromatin.
Dong W, Oya E, Zahedi Y, Prasad P, Svensson JP, Lennartsson A, Ekwall K, Durand-Dubief M
Heterochromatin regulation is critical for genomic stability. Different H3K9 methylation states have been discovered, with distinct roles in heterochromatin formation and silencing. However, how the transition from H3K9me2 to H3K9me3 is controlled is still unclear. Here, we investigate the role of the conserved brom... |
Native Chromatin Proteomics Reveals a Role for Specific Nucleoporins in Heterochromatin Organization and Maintenance.
Iglesias N, Paulo JA, Tatarakis A, Wang X, Edwards AL, Bhanu NV, Garcia BA, Haas W, Gygi SP, Moazed D
Spatially and functionally distinct domains of heterochromatin and euchromatin play important roles in the maintenance of chromosome stability and regulation of gene expression, but a comprehensive knowledge of their composition is lacking. Here, we develop a strategy for the isolation of native Schizosaccharom... |
Replication timing and epigenome remodelling are associated with the nature of chromosomal rearrangements in cancer.
Du Q, Bert SA, Armstrong NJ, Caldon CE, Song JZ, Nair SS, Gould CM, Luu PL, Peters T, Khoury A, Qu W, Zotenko E, Stirzaker C, Clark SJ
DNA replication timing is known to facilitate the establishment of the epigenome, however, the intimate connection between replication timing and changes to the genome and epigenome in cancer remain largely uncharacterised. Here, we perform Repli-Seq and integrated epigenome analyses and demonstrate that genomic reg... |
Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability.
Iglesias N, Currie MA, Jih G, Paulo JA, Siuti N, Kalocsay M, Gygi SP, Moazed D
Histone H3 lysine 9 methylation (H3K9me) mediates heterochromatic gene silencing and is important for genome stability and the regulation of gene expression. The establishment and epigenetic maintenance of heterochromatin involve the recruitment of H3K9 methyltransferases to specific sites on DNA, followed by the re... |
Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation.
Yu R, Wang X, Moazed D
Histone post-translational modifications (PTMs) are associated with epigenetic states that form the basis for cell-type-specific gene expression. Once established, histone PTMs can be maintained by positive feedback involving enzymes that recognize a pre-existing histone modification and catalyse the same modificati... |
Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription
Jih G. et al.
Heterochromatic DNA domains have important roles in the regulation of gene expression and maintenance of genome stability by silencing repetitive DNA elements and transposons. From fission yeast to mammals, heterochromatin assembly at DNA repeats involves the activity of small noncoding RNAs (sRNAs) associated with ... |