Notice (8): Undefined variable: solution_of_interest [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'carrousel_order' => array(
(int) 0 => 'ChIPmentation'
),
'language' => 'cn',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'meta_title' => 'Auto True MicroChIP kit',
'product' => array(
'Product' => array(
'id' => '1858',
'antibody_id' => null,
'name' => 'Auto True MicroChIP kit',
'description' => '<p>The Auto True MicroChIP kit in combination with the <a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns">MicroPlex Library Preparation™ kit</a> allows for performing ChIP-seq on as few as 10,000 cells. This microChIP-seq assay has been validated with the new generation <a href="../p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star<sup>®</sup> Compact Automated Workstation</a>. Furthermore Diagenode's high quality ChIP-seq grade antibodies have been used.</p>
<div class="row">
<div class="small-12 medium-5 large-6 columns"><small style="color: #666;">Video article</small>
<div class="flex-video"><iframe width="420" height="315" style="float: left; margin-right: 20px;" src="//www.youtube.com/embed/JZC9ARbLh_U?rel=0" frameborder="0" allowfullscreen="allowfullscreen"></iframe></div>
</div>
<div class="small-12 medium-7 large-6 columns">
<h3>Automating ChIP-seq Experiments to Generate Epigenetic Profiles on 10,000 HeLa Cells</h3>
<p><a href="https://www.youtube.com/watch?v=JZC9ARbLh_U"><img src="https://www.diagenode.com/img/jove_logo.png" alt="Jove logo" border="0" width="100" height="58" /></a></p>
</div>
</div>',
'label1' => '',
'info1' => '',
'label2' => 'Characteristics',
'info2' => '<ul>
<li><strong>Revolutionary:</strong> Only 10.000 cells needed for complete ChIP-seq procedure</li>
<li><strong>Reliable and accurate</strong> sequencing library preparation from just picogram inputs</li>
<li><strong>Validated on</strong> studies for histone marks and with the IP-Star<sup>®</sup> Compact Automated Platform</li>
</ul>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-histone-results.png" alt="ChIP on 10,000 cells" style="display: block; margin-left: auto; margin-right: auto;" /></p>
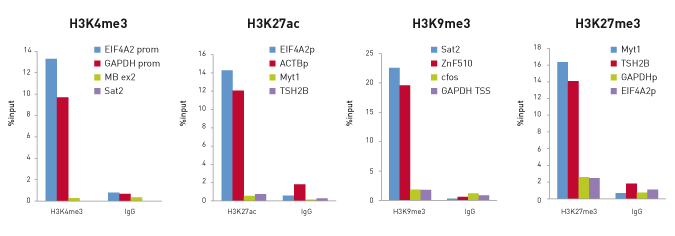
<p><strong>High efficiency ChIP on 10,000 cells</strong><br />ChIP efficiency on 10 000 cells. ChIP was performed on human Hela cells using the Diagenode antibodies H3K4me3 (Cat. No. pAb-003-050), H3K27ac (pAb-174-050), H3K9me3 (pAb-056-050) and H3K27me3 (pAb-069-050). Sheared chromatin from 10 000 cells and 0.1 µg (H3K27ac), 0.25 µg (H3K4me3 and H3K27me3) or 0.5 µg (H3K9me3) of the antibody were used per IP. Corresponding amount of IgG was used as control. Quantitative PCR was performed with primers for corresponding positive and negative loci. Figure shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-A.png" alt="ChIP amplification" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-B.png" alt="Match between True MicroChIP peaks and the reference dataset" style="display: block; margin-left: auto; margin-right: auto;" /></p>
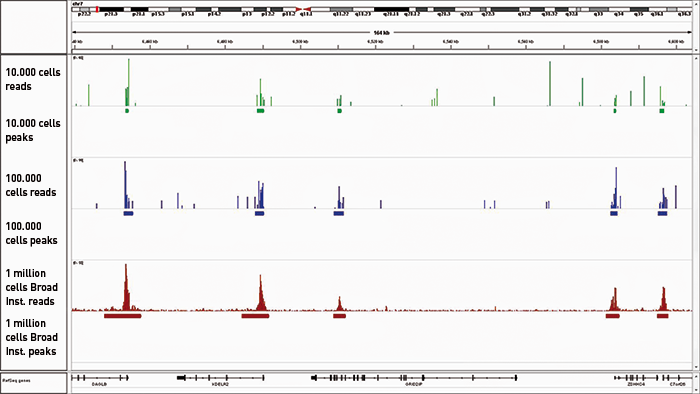
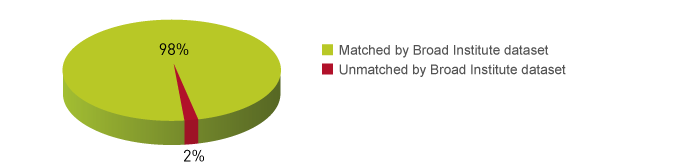
<p><strong>Reliable detection of enrichments in ChIP-seq</strong><br />A: ChIP has been peformed with H3K4me3 antibody, amplification of 17 pg of DNA ChIP'd from 10.000 cells and amplification of 35 pg of DNA ChIP'd from 100.000 cells (control experiment). The IP'd DNA was amplified and transformed into a sequencing-ready preparation for the Illumina plateform with the MicroPlex Library Preparation kit. The library was then analysed on an Illumina<sup>®</sup> Genome Analyzer. Cluster generation and sequencing were performed according to the manufacturer's instructions.<br />B: We observed a perfect match between the top 40% of True MicroChIP peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-result.png" alt="Cell lysis" style="display: block; margin-left: auto; margin-right: auto;" /></p>
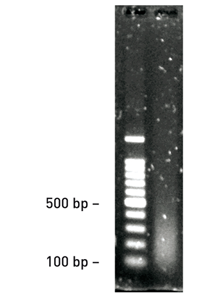
<p><strong>Hela cells were fixed with 1% formaldehyde (for 10 minutes at RT)</strong><br /> Cell lysis was performed using the Lysis Buffer tL1 of the Diagenode True MicroChIP kit. Samples corresponding to 10 000 cells are sheared during 5 rounds of 5 cycles of 30 seconds “ON” / 30 seconds “OFF” with the Bioruptor<sup>®</sup> Plus combined with the Bioruptor<sup>®</sup> Water cooler (Cat No. BioAcc-cool) at HIGH power setting (position H). For optimal results, samples are vortexed before and after performing 5 sonication cycles, followed by a short centrifugation at 4°C. 10 μl of DNA (equivalent to 60 000 cells) are analysed on a 1.5% agarose gel.</p>',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C01010140',
'old_catalog_number' => '',
'sf_code' => 'C01010140-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '540',
'price_USD' => '590',
'price_GBP' => '495',
'price_JPY' => '84590',
'price_CNY' => '',
'price_AUD' => '1475',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-true-microchip-kit-16-rxns',
'meta_title' => 'Auto True MicroChIP kit',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'modified' => '2019-05-10 09:28:26',
'created' => '2015-06-29 14:08:20',
'locale' => 'zho'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(),
'Group' => array(),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array()
)
)
$carrousel_order = array(
(int) 0 => 'ChIPmentation'
)
$language = 'cn'
$meta_keywords = ''
$meta_description = 'Auto True MicroChIP kit'
$meta_title = 'Auto True MicroChIP kit'
$product = array(
'Product' => array(
'id' => '1858',
'antibody_id' => null,
'name' => 'Auto True MicroChIP kit',
'description' => '<p>The Auto True MicroChIP kit in combination with the <a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns">MicroPlex Library Preparation™ kit</a> allows for performing ChIP-seq on as few as 10,000 cells. This microChIP-seq assay has been validated with the new generation <a href="../p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star<sup>®</sup> Compact Automated Workstation</a>. Furthermore Diagenode's high quality ChIP-seq grade antibodies have been used.</p>
<div class="row">
<div class="small-12 medium-5 large-6 columns"><small style="color: #666;">Video article</small>
<div class="flex-video"><iframe width="420" height="315" style="float: left; margin-right: 20px;" src="//www.youtube.com/embed/JZC9ARbLh_U?rel=0" frameborder="0" allowfullscreen="allowfullscreen"></iframe></div>
</div>
<div class="small-12 medium-7 large-6 columns">
<h3>Automating ChIP-seq Experiments to Generate Epigenetic Profiles on 10,000 HeLa Cells</h3>
<p><a href="https://www.youtube.com/watch?v=JZC9ARbLh_U"><img src="https://www.diagenode.com/img/jove_logo.png" alt="Jove logo" border="0" width="100" height="58" /></a></p>
</div>
</div>',
'label1' => '',
'info1' => '',
'label2' => 'Characteristics',
'info2' => '<ul>
<li><strong>Revolutionary:</strong> Only 10.000 cells needed for complete ChIP-seq procedure</li>
<li><strong>Reliable and accurate</strong> sequencing library preparation from just picogram inputs</li>
<li><strong>Validated on</strong> studies for histone marks and with the IP-Star<sup>®</sup> Compact Automated Platform</li>
</ul>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-histone-results.png" alt="ChIP on 10,000 cells" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>High efficiency ChIP on 10,000 cells</strong><br />ChIP efficiency on 10 000 cells. ChIP was performed on human Hela cells using the Diagenode antibodies H3K4me3 (Cat. No. pAb-003-050), H3K27ac (pAb-174-050), H3K9me3 (pAb-056-050) and H3K27me3 (pAb-069-050). Sheared chromatin from 10 000 cells and 0.1 µg (H3K27ac), 0.25 µg (H3K4me3 and H3K27me3) or 0.5 µg (H3K9me3) of the antibody were used per IP. Corresponding amount of IgG was used as control. Quantitative PCR was performed with primers for corresponding positive and negative loci. Figure shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-A.png" alt="ChIP amplification" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-B.png" alt="Match between True MicroChIP peaks and the reference dataset" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Reliable detection of enrichments in ChIP-seq</strong><br />A: ChIP has been peformed with H3K4me3 antibody, amplification of 17 pg of DNA ChIP'd from 10.000 cells and amplification of 35 pg of DNA ChIP'd from 100.000 cells (control experiment). The IP'd DNA was amplified and transformed into a sequencing-ready preparation for the Illumina plateform with the MicroPlex Library Preparation kit. The library was then analysed on an Illumina<sup>®</sup> Genome Analyzer. Cluster generation and sequencing were performed according to the manufacturer's instructions.<br />B: We observed a perfect match between the top 40% of True MicroChIP peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-result.png" alt="Cell lysis" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Hela cells were fixed with 1% formaldehyde (for 10 minutes at RT)</strong><br /> Cell lysis was performed using the Lysis Buffer tL1 of the Diagenode True MicroChIP kit. Samples corresponding to 10 000 cells are sheared during 5 rounds of 5 cycles of 30 seconds “ON” / 30 seconds “OFF” with the Bioruptor<sup>®</sup> Plus combined with the Bioruptor<sup>®</sup> Water cooler (Cat No. BioAcc-cool) at HIGH power setting (position H). For optimal results, samples are vortexed before and after performing 5 sonication cycles, followed by a short centrifugation at 4°C. 10 μl of DNA (equivalent to 60 000 cells) are analysed on a 1.5% agarose gel.</p>',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C01010140',
'old_catalog_number' => '',
'sf_code' => 'C01010140-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '540',
'price_USD' => '590',
'price_GBP' => '495',
'price_JPY' => '84590',
'price_CNY' => '',
'price_AUD' => '1475',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-true-microchip-kit-16-rxns',
'meta_title' => 'Auto True MicroChIP kit',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'modified' => '2019-05-10 09:28:26',
'created' => '2015-06-29 14:08:20',
'locale' => 'zho'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(),
'Group' => array(),
'Related' => array(
(int) 0 => array(
'id' => '1885',
'antibody_id' => null,
'name' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'description' => '<p><span>The Auro hMeDIP kit is designed for enrichment of hydroxymethylated DNA from fragmented genomic DNA samples for use in genome-wide methylation analysis. It features</span><span> a highly specific monoclonal antibody against </span><span>5-hydroxymethylcytosine (5-hmC) for the immunoprecipitation of hydroxymethylated DNA</span><span>. It includes control DNA and primers to assess the effiency of the assay. </span><span>Performing hydroxymethylation profiling with the hMeDIP kit is fast, reliable and highly specific.</span></p>',
'label1' => ' Characteristics',
'info1' => '<ul style="list-style-type: disc;">
<li><span>Robust enrichment & immunoprecipitation of hydroxymethylated DNA</span></li>
<li>Highly specific monoclonal antibody against 5-hmC<span> for reliable, reproducible results</span></li>
<li>Including control DNA and primers to <span>monitor the efficiency of the assay</span>
<ul style="list-style-type: circle;">
<li>5-hmC, 5-mC and unmethylated DNA sequences and primer pairs</li>
<li>Mouse primer pairs against Sfi1 targeting hydroxymethylated gene in mouse</li>
</ul>
</li>
</ul>
<ul style="list-style-type: disc;">
<li>Improved single-tube, magnetic bead-based protocol</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C02010034',
'old_catalog_number' => '',
'sf_code' => 'C02010034-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '630',
'price_USD' => '690',
'price_GBP' => '580',
'price_JPY' => '98690',
'price_CNY' => '',
'price_AUD' => '1725',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns',
'meta_title' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'meta_keywords' => '',
'meta_description' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'modified' => '2021-01-18 10:37:19',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1893',
'antibody_id' => null,
'name' => 'Auto Premium Bisulfite kit',
'description' => '<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => 'Characteristics',
'info1' => '<p><strong>Bisulfite Conversion efficiency in TERTBS1 and CTS56 genomic regions.</strong></p>
<p>The figure shows the conversion efficiency in TERTBS1 and CTS56 genomic regions when using 1 μg, 500ng and 100 ng of genomic DNA. Between 1 and 5 clones were analyzed for the different amounts of genomic DNA</p>
<p><img src="https://www.diagenode.com/img/product/kits/auto-premium-bisulfite-clone.png" alt="Clone" /></p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '40 rxns',
'catalog_number' => 'C02030031',
'old_catalog_number' => '',
'sf_code' => 'C02030031-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-premium-bisulfite-kit-40-rxns',
'meta_title' => 'Auto Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Auto Premium Bisulfite kit',
'modified' => '2019-12-16 10:05:26',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1842',
'antibody_id' => null,
'name' => 'Auto iDeal ChIP-seq Kit for Transcription Factors',
'description' => '<p><span><strong>This product must be used with the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star Compact Automated System</a>.</strong></span></p>
<p><span>Diagenode’s </span><strong>Auto iDeal ChIP-seq Kit for Transcription Factors</strong><span> is a highly specialized solution for robust Transcription Factor ChIP-seq results. Unlike competing solutions, our kit utilizes a highly optimized protocol and is backed by validation with a broad number and range of transcription factors. The kit provides high yields with excellent specificity and sensitivity.</span></p>',
'label1' => 'Characteristics',
'info1' => '<ul>
<li><strong>Confidence in results:</strong> Validated for ChIP-seq with multiple transcription factors</li>
<li><strong>Proven:</strong> Validated by the epigenetics community, including the BLUEPRINT consortium</li>
<li><strong>Most complete kit available</strong> for highest quality data - includes control antibodies and primers</li>
<li>Validated with Diagenode's <a href="https://www.diagenode.com/en/p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns"><span>MicroPlex Library Preparation™ kit</span></a> and <a href="https://www.diagenode.com/categories/ip-star" title="IP-Star Automated System">IP-Star<sup>®</sup></a> Automation System</li>
</ul>
<p> </p>
<h3>ChIP-seq on cells</h3>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-ctcf-diagenode.jpg" alt="CTCF Diagenode" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1.</strong> (A) Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina<sup>®</sup> HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the GAPDH positive control gene.</p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-figure-b-total-diagendoe-peaks.png" alt="CTCF Diagenode" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1B.</strong> The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p> </p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-A.png" alt="ChIP-seq figure A" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-B.png" alt="ChIP-seq figure B" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-C.png" alt="ChIP-seq figure C" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 2.</strong> Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade HDAC1 (A), LSD1 (B) and p53 antibody (C). The IP'd DNA was subsequently analysed on an Illumina<sup>®</sup> Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in regions of chromosome 3 (A), chromosome 12 (B) and chromosome 6 (C) respectively.</p>
<p> </p>
<h3>ChIP-seq on tissue</h3>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-figure-3a.jpg" alt="ChIP-seq figure A" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 3A.</strong> Chromatin Immunoprecipitation has been performed using chromatin from mouse liver tissue, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina® HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the Vwf positive control gene.</p>
<p><img src="https://www.diagenode.com/img/product/kits/match-of-the-top40-peaks.png" alt="Match of the Top40 peaks" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 3B.</strong> The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>',
'label2' => 'Species, cell lines, tissues tested',
'info2' => '<p>The iDeal ChIP-seq Kit for Transcription Factors is compatible with a broad variety of cell lines, tissues and species, as shown below. Other species / cell lines / tissues can be used with this kit.</p>
<p><span style="text-decoration: underline;">Cell lines:</span></p>
<p>Human: A549, A673, BT-549, CD4 T, HCC1806, HeLa, HepG2, HFF, HK-GFP-MR, ILC, K562, KYSE-180, LapC4, M14, MCF7, MDA-MB-231, MDA-MB-436, RDES, SKNO1, VCaP, U2-OS, ZR-75-1 </p>
<p>Mouse: ESC, NPCs, BZ, GT1-7, acinar cells, HSPCs, Th2 cells, keratinocytes</p>
<p>Cattle: pbMEC, <span>MAC-T</span></p>
<p><span style="text-decoration: underline;">Tissues:</span></p>
<p>Mouse: kidney, heart, brain, iris, liver, limbs from E10.5 embryos</p>
<p><span>Horse: l</span>iver, brain, heart, lung, skeletal muscle, lamina, ovary</p>
<p><span style="text-decoration: underline;">ChIP on yeast</span></p>
<p>The iDeal ChIP-seq kit for TF is compatible with yeast samples. Check out our <strong><a href="https://www.diagenode.com/files/products/kits/Application_Note-ChIP_on_Yeast.pdf">Application Note</a></strong> presenting an optimized detailed protocol for ChIP on yeast.</p>
<p></p>
<p>Did you use the iDeal ChIP-seq for Transcription Factors Kit on other cell line / tissue / species? <a href="mailto:agnieszka.zelisko@diagenode.com?subject=Species, cell lines, tissues tested with the iDeal ChIP-seq Kit for TF&body=Dear Customer,%0D%0A%0D%0APlease, leave below your feedback about the iDeal ChIP-seq for Transcription Factors (cell / tissue type, species, other information...).%0D%0A%0D%0AThank you for sharing with us your experience !%0D%0A%0D%0ABest regards,%0D%0A%0D%0AAgnieszka Zelisko-Schmidt, PhD">Let us know!</a></p>',
'label3' => 'Additional solutions compatible with Auto iDeal ChIP-seq kit for Transcription Factors',
'info3' => '<p><span style="font-weight: 400;">The</span> <a href="https://www.diagenode.com/en/p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns"><span style="font-weight: 400;">Chromatin shearing optimization kit – Low SDS (iDeal Kit for TFs)</span></a><span style="font-weight: 400;"> is the kit compatible with the iDeal ChIP-seq kit for TF, recommended for the optimization of chromatin shearing, a critical step for ChIP.</span></p>
<p><a href="https://www.diagenode.com/en/p/chip-cross-link-gold-600-ul"><span style="font-weight: 400;">ChIP Cross-link Gold</span></a> <span style="font-weight: 400;">should be used in combination with formaldehyde when working with higher order and/or dynamic interactions, for efficient protein-protein fixation.</span></p>
<p><span style="font-weight: 400;">For library preparation of immunoprecipitated samples we recommend to use the </span><b> </b><a href="https://www.diagenode.com/en/categories/library-preparation-for-ChIP-seq"><span style="font-weight: 400;">MicroPlex Library Preparation Kit</span></a><span style="font-weight: 400;"> - validated for library preparation from picogram inputs.</span></p>
<p><a href="https://www.diagenode.com/en/categories/chip-seq-grade-antibodies"><span style="font-weight: 400;">ChIP-seq grade antibodies</span></a><span style="font-weight: 400;"> provide high yields with excellent specificity and sensitivity.</span></p>
<p><span style="font-weight: 400;">Check the list of available </span><a href="https://www.diagenode.com/en/categories/primer-pairs"><span style="font-weight: 400;">Primer pairs</span></a><span style="font-weight: 400;"> designed for high specificity to specific genomic regions.</span></p>',
'format' => '24 rxns',
'catalog_number' => 'C01010058',
'old_catalog_number' => '',
'sf_code' => 'C01010058-',
'type' => 'RFR',
'search_order' => '01-Accessory',
'price_EUR' => '915',
'price_USD' => '1130',
'price_GBP' => '840',
'price_JPY' => '143335',
'price_CNY' => '',
'price_AUD' => '2825',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns',
'meta_title' => 'Auto iDeal ChIP-seq Kit for Transcription Factors x24',
'meta_keywords' => '',
'meta_description' => 'Auto iDeal ChIP-seq Kit for Transcription Factors x24',
'modified' => '2021-11-23 10:51:46',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1923',
'antibody_id' => null,
'name' => 'MicroChIP DiaPure columns',
'description' => '<p><a href="https://www.diagenode.com/files/products/reagents/MicroChIP_DiaPure_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p>Diagenode’s MicroChIP DiaPure columns have been optimized for the purification and elution of very low amounts of DNA. This rapid method has been validated for epigenetic applications like low input ChIP (e.g. using the True MicroChIP kit) and CUT&Tag (e.g. using Diagenode’s pA-Tn5), but is also compatible with many other applications. The DNA can be eluted at high concentrations in volumes down to 6 μl and it is suitable for any downstream application (e.g. NGS).</p>
<p>Benefits of the MicroChIP DiaPure columns:</p>
<ul>
<li>Optimized for the purification of very low DNA amounts</li>
<li>Fast and easy protocol</li>
<li>Non-toxic</li>
<li>Validated for ChIP and Cut&Tag</li>
</ul>',
'label1' => 'Examples of results',
'info1' => '<h2 style="text-align: center;">MicroChIP DiaPure columns after ChIP</h2>
<p>Successful ChIP-seq results generated on 50,000 of K562 cells using True MicroChIP technology. ChIP has been performed accordingly to True MicroChIP protocol (Diagenode, Cat. No. C01010130), including DNA purification using the MicroChIP DiaPure columns. For the library preparation the MicroPlex Library Preparation Kit (Diagenode, Cat. No. C05010001) has been used. The below figure shows the peaks from ChIP-seq experiments using the following Diagenode antibodies: H3K4me1 (C15410194), H3K9/14ac (C15410200), H3K27ac (C15410196) and H3K36me3 (C15410192).</p>
<p><img src="https://www.diagenode.com/img/product/kits/figure-igv-microchip.png" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1:</strong> Integrative genomics viewer (IGV) visualization of ChIP-seq experiments using 50,000 of K562 cells.</p>
<p></p>
<h2 style="text-align: center;"></h2>
<h2 style="text-align: center;">MicroChIP DiaPure columns after CUT&Tag</h2>
<p>Successful CUT&Tag results showing a low background with high region-specific enrichment has been generated using 50.000 of K562 cells, 1 µg of H3K27me3 antibody (Diagenode, Cat. No. C15410069) and proteinA-Tn5 (1:250) (Diagenode, Cat. No. C01070001). 1 µg of IgG (Diagenode, Cat. No. C15410206) was used as negative control. Samples were purified using the MicroChIP DiaPure columns or phenol-chloroform purification. The below figure presenst the comparison of two purification methods.</p>
<p><img src="https://www.diagenode.com/img/product/kits/figure-diapure-igv.png" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 2:</strong> Integrative genomics viewer (IGV) visualization of CUT&Tag experiments: MicroChIP DiaPure columns vs phenol-chloroform purification using the H3K27me3 antibody.</p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C03040001',
'old_catalog_number' => '',
'sf_code' => 'C03040001-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'microchip-diapure-columns-50-rxns',
'meta_title' => 'MicroChIP DiaPure columns',
'meta_keywords' => '',
'meta_description' => 'MicroChIP DiaPure columns',
'modified' => '2023-04-20 16:11:26',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array([maximum depth reached])
),
(int) 4 => array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '9',
'position' => '10',
'parent_id' => '2',
'name' => 'ChIP-seq',
'description' => '<div class="row">
<div class="large-12 columns">Chromatin Immunoprecipitation (ChIP) coupled with high-throughput massively parallel sequencing as a detection method (ChIP-seq) has become one of the primary methods for epigenomics researchers, namely to investigate protein-DNA interaction on a genome-wide scale. This technique is now used in a variety of life science disciplines including cellular differentiation, tumor suppressor gene silencing, and the effect of histone modifications on gene expression.</div>
<div class="large-12 columns"></div>
<h5 class="large-12 columns"><strong></strong></h5>
<h5 class="large-12 columns"><strong>The ChIP-seq workflow</strong></h5>
<div class="small-12 medium-12 large-12 columns text-center"><br /><img src="https://www.diagenode.com/img/chip-seq-diagram.png" /></div>
<div class="large-12 columns"><br />
<ol>
<li class="large-12 columns"><strong>Chromatin preparation: </strong>Crosslink chromatin-bound proteins (histones or transcription factors) to DNA followed by cell lysis.</li>
<li class="large-12 columns"><strong>Chromatin shearing:</strong> Fragment chromatin by sonication to desired fragment size (100-500 bp)</li>
<li class="large-12 columns"><strong>Chromatin IP</strong>: Capture protein-DNA complexes with <strong><a href="../categories/chip-seq-grade-antibodies">specific ChIP-seq grade antibodies</a></strong> against the histone or transcription factor of interest</li>
<li class="large-12 columns"><strong>DNA purification</strong>: Reverse cross-links, elute, and purify </li>
<li class="large-12 columns"><strong>NGS Library Preparation</strong>: Ligate adapters and amplify IP'd material</li>
<li class="large-12 columns"><strong>Bioinformatic analysis</strong>: Perform r<span style="font-weight: 400;">ead filtering and trimming</span>, r<span style="font-weight: 400;">ead specific alignment, enrichment specific peak calling, QC metrics, multi-sample cross-comparison etc. </span></li>
</ol>
</div>
</div>
<div class="row" style="margin-top: 32px;">
<div class="small-12 medium-10 large-9 small-centered columns">
<div class="radius panel" style="background-color: #fff;">
<h3 class="text-center" style="color: #b21329;">Need guidance?</h3>
<p class="text-justify">Choose our full ChIP kits or simply choose what you need from antibodies, buffers, beads, chromatin shearing and purification reagents. With the ChIP Kit Customizer, you have complete flexibility on which components you want from our validated ChIP kits.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns"><a href="../pages/which-kit-to-choose"><img alt="" src="https://www.diagenode.com/img/banners/banner-decide.png" /></a></div>
<div class="small-6 medium-6 large-6 columns"><a href="../pages/chip-kit-customizer-1"><img alt="" src="https://www.diagenode.com/img/banners/banner-customizer.png" /></a></div>
</div>
</div>
</div>
</div>',
'in_footer' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'slug' => 'chromatin-immunoprecipitation-sequencing',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers wide range of kits and antibodies for Chromatin Immunoprecipitation Sequencing (ChIP-Seq) and also provides Bioruptor for chromatin shearing',
'meta_title' => 'Chromatin Immunoprecipitation - ChIP-seq Kits - Dna methylation | Diagenode',
'modified' => '2017-11-14 09:57:16',
'created' => '2015-04-12 18:08:46',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '117',
'position' => '3',
'parent_id' => '59',
'name' => 'Automated kits',
'description' => '',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'hide' => false,
'all_format' => false,
'is_antibody' => false,
'slug' => 'chromatin-ip-automated-kits',
'cookies_tag_id' => null,
'meta_keywords' => '',
'meta_description' => 'Diagenode’s validated ChIP-Seq Automated kits has everything you need for a successful ChIP prior to Next-Generation Sequencing.',
'meta_title' => 'Automated ChIP-Seq Kits for Chromatin Immunoprecipitation | Diagenode',
'modified' => '2022-02-11 14:11:01',
'created' => '2016-07-19 16:57:20',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '110',
'name' => 'Auto TrueMicroChIP kit',
'description' => '<div class="page" title="Page 4">
<div class="layoutArea">
<div class="column">
<p><span></span><span>Conventional ChIP protocols require high numbers of cells (hundreds of thousands cells at least) limiting the application for ChIP technology to few cell samples. Recently, ChIP assays on smallest amount of cells have been reported. The procedure requires tedious optimization of several reaction conditions to face the increased background observed in ChIP performed with reduced amount of cells. That might consequently lead to considerable time and lab expenditures. To reduce these tedious steps, Diagenode provides the new Auto True MicroChIP kit with optimized reagents and protocol to enable successful ChIP on as few as 10 000 cells. Moreover, the Auto True MicroChIP kit protocol has been thoroughly optimised by Diagenode for ChIP followed by high-throughput sequencing on Illumina</span><span>® </span><span>Next-Gen sequencers. To allow the generation of consistent results in ChIP-seq on 10 000 cells, Diagenode has also optimised a library preparation protocol on limited amount of immunoprecipitated DNA. </span></p>
<p><span>The new MicroPLEX library preparation kit allows the preparation of libraries for sequencing on picogram amount of immunoprecipitated DNA. Thus, association of the Auto True MicroChIP kit with the MicroPLEX library preparation kit provides optimised solutions to perform ChIP-sequencing on limited amount of cells. </span></p>
</div>
</div>
</div>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/Auto-TrueMicroChIP-kit-manual.pdf',
'slug' => 'auto-truemicrochip-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-09-01 12:30:58',
'created' => '2015-07-07 11:47:43',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '37',
'name' => 'Chromatin Brochure',
'description' => '<p>Whether you are experienced or new to the field of chromatin immunoprecipitation, Diagenode has everything you need to make ChIP easy and convenient while ensuring consistent data between samples and experiments. As an expert in the field of epigenetics, Diagenode is committed to providing complete solutions from chromatin shearing reagents, shearing instruments such as the Bioruptor® (the gold standard for chromatin shearing), ChIP kits, the largest number of validated and trusted antibodies on the market, and the SX-8G IP-Star® Compact Automated System to achieve unparalleled productivity and reproducibility.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf',
'slug' => 'chromatin-immunoprecipitation-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2022-03-24 12:34:11',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1775',
'name' => 'product/kits/chip-kit-icon.png',
'alt' => 'ChIP kit icon',
'modified' => '2018-04-17 11:52:29',
'created' => '2018-03-15 15:50:34',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '3563',
'name' => 'EZH2 is overexpressed in transitional preplasmablasts and is involved in human plasma cell differentiation.',
'authors' => 'Herviou L, Jourdan M, Martinez AM, Cavalli G, Moreaux J',
'description' => '<p>Plasma cells (PCs) play a major role in the defense of the host organism against pathogens. We have shown that PC generation can be modeled using multi-step culture systems that reproduce the sequential cell differentiation occurring in vivo. Using this unique model, we investigated the role of EZH2 during PC differentiation (PCD) using H3K27me3 and EZH2 ChIP-binding profiles. We then studied the effect of the inhibition of EZH2 enzymatic activity to understand how EZH2 regulates the key functions involved in PCD. EZH2 expression significantly increases in preplasmablasts with H3K27me3 mediated repression of genes involved in B cell and plasma cell identity. EZH2 was also found to be recruited to H3K27me3-free promoters of transcriptionally active genes known to regulate cell proliferation. Inhibition the catalytic activity of EZH2 resulted in B to PC transcriptional changes associated with PC maturation induction, as well as higher immunoglobulin secretion. Altogether, our data suggest that EZH2 is involved in the maintenance of preplasmablast transitory immature proliferative state that supports their amplification.</p>',
'date' => '2019-02-12',
'pmid' => 'http://www.pubmed.gov/30755708',
'doi' => '10.1038/s41375-019-0392-1',
'modified' => '2019-03-21 17:17:48',
'created' => '2019-03-21 14:12:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '3389',
'name' => 'An Integrated and Semiautomated Microscaled Approach to Profile Cis-Regulatory Elements by Histone Modification ChIP-Seq for Large-Scale Epigenetic Studies.',
'authors' => 'Youhanna Jankeel D, Cayford J, Schmiedel BJ, Vijayanand P, Seumois G.',
'description' => '<p>Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) is the preferred approach to map histone modifications and identify cis-regulatory DNA elements throughout the genome. Multiple methods have been described to increase the efficiency of library preparation and to reduce hands-on time as well as costs. This review describes detailed steps to perform cell fixation, chromatin shearing, immunoprecipitation, and sequencing library preparation for a batch of 48-96 samples with small cell numbers. The protocol implements a semiautomated platform to reduce technical variability and improve signal-to-noise ratio as well as reduce hands-on time, thus allowing large-scale epigenetic studies of clinical samples with limited cell numbers.</p>',
'date' => '2018-06-30',
'pmid' => 'http://www.pubmed.gov/29956160',
'doi' => '10.1007/978-1-4939-7896-0_22',
'modified' => '2018-11-09 12:29:33',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '3595',
'name' => 'Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche.',
'authors' => 'Baghdadi MB, Castel D, Machado L, Fukada SI, Birk DE, Relaix F, Tajbakhsh S, Mourikis P',
'description' => '<p>The cell microenvironment, which is critical for stem cell maintenance, contains both cellular and non-cellular components, including secreted growth factors and the extracellular matrix. Although Notch and other signalling pathways have previously been reported to regulate quiescence of stem cells, the composition and source of molecules that maintain the stem cell niche remain largely unknown. Here we show that adult muscle satellite (stem) cells in mice produce extracellular matrix collagens to maintain quiescence in a cell-autonomous manner. Using chromatin immunoprecipitation followed by sequencing, we identified NOTCH1/RBPJ-bound regulatory elements adjacent to specific collagen genes, the expression of which is deregulated in Notch-mutant mice. Moreover, we show that Collagen V (COLV) produced by satellite cells is a critical component of the quiescent niche, as depletion of COLV by conditional deletion of the Col5a1 gene leads to anomalous cell cycle entry and gradual diminution of the stem cell pool. Notably, the interaction of COLV with satellite cells is mediated by the Calcitonin receptor, for which COLV acts as a surrogate local ligand. Systemic administration of a calcitonin derivative is sufficient to rescue the quiescence and self-renewal defects found in COLV-null satellite cells. This study reveals a Notch-COLV-Calcitonin receptor signalling cascade that maintains satellite cells in a quiescent state in a cell-autonomous fashion, and raises the possibility that similar reciprocal mechanisms act in diverse stem cell populations.</p>',
'date' => '2018-05-23',
'pmid' => 'http://www.pubmed.gov/29795344',
'doi' => '10.1038/s41586-018-0144-9',
'modified' => '2019-04-17 15:12:55',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '3589',
'name' => 'A new metabolic gene signature in prostate cancer regulated by JMJD3 and EZH2.',
'authors' => 'Daures M, Idrissou M, Judes G, Rifaï K, Penault-Llorca F, Bignon YJ, Guy L, Bernard-Gallon D',
'description' => '<p>Histone methylation is essential for gene expression control. Trimethylated lysine 27 of histone 3 (H3K27me3) is controlled by the balance between the activities of JMJD3 demethylase and EZH2 methyltransferase. This epigenetic mark has been shown to be deregulated in prostate cancer, and evidence shows H3K27me3 enrichment on gene promoters in prostate cancer. To study the impact of this enrichment, a transcriptomic analysis with TaqMan Low Density Array (TLDA) of several genes was studied on prostate biopsies divided into three clinical grades: normal ( = 23) and two tumor groups that differed in their aggressiveness (Gleason score ≤ 7 ( = 20) and >7 ( = 19)). ANOVA demonstrated that expression of the gene set was upregulated in tumors and correlated with Gleason score, thus discriminating between the three clinical groups. Six genes involved in key cellular processes stood out: , , , , and . Chromatin immunoprecipitation demonstrated collocation of EZH2 and JMJD3 on gene promoters that was dependent on disease stage. Gene set expression was also evaluated on prostate cancer cell lines (DU 145, PC-3 and LNCaP) treated with an inhibitor of JMJD3 (GSK-J4) or EZH2 (DZNeP) to study their involvement in gene regulation. Results showed a difference in GSK-J4 sensitivity under PTEN status of cell lines and an opposite gene expression profile according to androgen status of cells. In summary, our data describe the impacts of JMJD3 and EZH2 on a new gene signature involved in prostate cancer that may help identify diagnostic and therapeutic targets in prostate cancer.</p>',
'date' => '2018-05-04',
'pmid' => 'http://www.pubmed.gov/29805743',
'doi' => '10.18632/oncotarget.25182',
'modified' => '2019-04-17 15:21:33',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '3193',
'name' => 'Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression',
'authors' => 'Ngollo M. et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">H3K27me3 histone marks shape the inhibition of gene transcription. In prostate cancer, the deregulation of H3K27me3 marks might play a role in prostate tumor progression.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">We investigated genome-wide H3K27me3 histone methylation profile using chromatin immunoprecipitation (ChIP) and 2X400K promoter microarrays to identify differentially-enriched regions in biopsy samples from prostate cancer patients. H3K27me3 marks were assessed in 34 prostate tumors: 11 with Gleason score > 7 (GS > 7), 10 with Gleason score ≤ 7 (GS ≤ 7), and 13 morphologically normal prostate samples.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">Here, H3K27me3 profiling identified an average of 386 enriched-genes on promoter regions in healthy control group versus 545 genes in GS ≤ 7 and 748 genes in GS > 7 group. We then ran a factorial discriminant analysis (FDA) and compared the enriched genes in prostate-tumor biopsies and normal biopsies using ANOVA to identify significantly differentially-enriched genes. The analysis identified ALG5, EXOSC8, CBX1, GRID2, GRIN3B, ING3, MYO1D, NPHP3-AS1, MSH6, FBXO11, SND1, SPATS2, TENM4 and TRA2A genes. These genes are possibly associated with prostate cancer. Notably, the H3K27me3 histone mark emerged as a novel regulatory mechanism in poor-prognosis prostate cancer.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Our findings point to epigenetic mark H3K27me3 as an important event in prostate carcinogenesis and progression. The results reported here provide new molecular insights into the pathogenesis of prostate cancer.</abstracttext></p>
</div>',
'date' => '2017-04-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28403887',
'doi' => '',
'modified' => '2017-06-19 14:07:35',
'created' => '2017-06-19 14:05:03',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array()
)
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array()
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010034</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1885" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1885" id="CartAdd/1885Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1885" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto hMeDIP kit x16 (monoclonal mouse antibody)</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns" data-reveal-id="cartModal-1885" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto hMeDIP kit x16 (monoclonal mouse antibody)</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-premium-bisulfite-kit-40-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02030031</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1893" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1893" id="CartAdd/1893Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1893" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto Premium Bisulfite kit</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto Premium Bisulfite kit',
'C02030031',
'240',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto Premium Bisulfite kit',
'C02030031',
'240',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-premium-bisulfite-kit-40-rxns" data-reveal-id="cartModal-1893" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto Premium Bisulfite kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns"><img src="/img/product/kits/chip-kit-icon.png" alt="ChIP kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C01010058</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1842" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1842" id="CartAdd/1842Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1842" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto iDeal ChIP-seq Kit for Transcription Factors</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto iDeal ChIP-seq Kit for Transcription Factors',
'C01010058',
'1130',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto iDeal ChIP-seq Kit for Transcription Factors',
'C01010058',
'1130',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns" data-reveal-id="cartModal-1842" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto iDeal ChIP-seq Kit for Transcription Factors</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/microchip-diapure-columns-50-rxns"><img src="/img/grey-logo.jpg" alt="default alt" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C03040001</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1923" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1923" id="CartAdd/1923Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1923" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MicroChIP DiaPure columns</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MicroChIP DiaPure columns',
'C03040001',
'125',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MicroChIP DiaPure columns',
'C03040001',
'125',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="microchip-diapure-columns-50-rxns" data-reveal-id="cartModal-1923" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MicroChIP DiaPure columns</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020011</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1888" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1888" id="CartAdd/1888Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1888" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto MethylCap kit</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1888" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto MethylCap kit</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '3624',
'product_id' => '1858',
'related_id' => '1888'
),
'Image' => array(
(int) 0 => array(
'id' => '1778',
'name' => 'product/kits/methyl-kit-icon.png',
'alt' => 'Methylation kit icon',
'modified' => '2019-04-23 15:17:01',
'created' => '2018-03-15 15:52:12',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$document = array(
'id' => '37',
'name' => 'Chromatin Brochure',
'description' => '<p>Whether you are experienced or new to the field of chromatin immunoprecipitation, Diagenode has everything you need to make ChIP easy and convenient while ensuring consistent data between samples and experiments. As an expert in the field of epigenetics, Diagenode is committed to providing complete solutions from chromatin shearing reagents, shearing instruments such as the Bioruptor® (the gold standard for chromatin shearing), ChIP kits, the largest number of validated and trusted antibodies on the market, and the SX-8G IP-Star® Compact Automated System to achieve unparalleled productivity and reproducibility.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf',
'slug' => 'chromatin-immunoprecipitation-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2022-03-24 12:34:11',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '1094',
'product_id' => '1858',
'document_id' => '37'
)
)
$publication = array(
'id' => '3193',
'name' => 'Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression',
'authors' => 'Ngollo M. et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">H3K27me3 histone marks shape the inhibition of gene transcription. In prostate cancer, the deregulation of H3K27me3 marks might play a role in prostate tumor progression.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">We investigated genome-wide H3K27me3 histone methylation profile using chromatin immunoprecipitation (ChIP) and 2X400K promoter microarrays to identify differentially-enriched regions in biopsy samples from prostate cancer patients. H3K27me3 marks were assessed in 34 prostate tumors: 11 with Gleason score > 7 (GS > 7), 10 with Gleason score ≤ 7 (GS ≤ 7), and 13 morphologically normal prostate samples.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">Here, H3K27me3 profiling identified an average of 386 enriched-genes on promoter regions in healthy control group versus 545 genes in GS ≤ 7 and 748 genes in GS > 7 group. We then ran a factorial discriminant analysis (FDA) and compared the enriched genes in prostate-tumor biopsies and normal biopsies using ANOVA to identify significantly differentially-enriched genes. The analysis identified ALG5, EXOSC8, CBX1, GRID2, GRIN3B, ING3, MYO1D, NPHP3-AS1, MSH6, FBXO11, SND1, SPATS2, TENM4 and TRA2A genes. These genes are possibly associated with prostate cancer. Notably, the H3K27me3 histone mark emerged as a novel regulatory mechanism in poor-prognosis prostate cancer.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Our findings point to epigenetic mark H3K27me3 as an important event in prostate carcinogenesis and progression. The results reported here provide new molecular insights into the pathogenesis of prostate cancer.</abstracttext></p>
</div>',
'date' => '2017-04-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28403887',
'doi' => '',
'modified' => '2017-06-19 14:07:35',
'created' => '2017-06-19 14:05:03',
'ProductsPublication' => array(
'id' => '2138',
'product_id' => '1858',
'publication_id' => '3193'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/28403887" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: header [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'carrousel_order' => array(
(int) 0 => 'ChIPmentation'
),
'language' => 'cn',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'meta_title' => 'Auto True MicroChIP kit',
'product' => array(
'Product' => array(
'id' => '1858',
'antibody_id' => null,
'name' => 'Auto True MicroChIP kit',
'description' => '<p>The Auto True MicroChIP kit in combination with the <a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns">MicroPlex Library Preparation™ kit</a> allows for performing ChIP-seq on as few as 10,000 cells. This microChIP-seq assay has been validated with the new generation <a href="../p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star<sup>®</sup> Compact Automated Workstation</a>. Furthermore Diagenode's high quality ChIP-seq grade antibodies have been used.</p>
<div class="row">
<div class="small-12 medium-5 large-6 columns"><small style="color: #666;">Video article</small>
<div class="flex-video"><iframe width="420" height="315" style="float: left; margin-right: 20px;" src="//www.youtube.com/embed/JZC9ARbLh_U?rel=0" frameborder="0" allowfullscreen="allowfullscreen"></iframe></div>
</div>
<div class="small-12 medium-7 large-6 columns">
<h3>Automating ChIP-seq Experiments to Generate Epigenetic Profiles on 10,000 HeLa Cells</h3>
<p><a href="https://www.youtube.com/watch?v=JZC9ARbLh_U"><img src="https://www.diagenode.com/img/jove_logo.png" alt="Jove logo" border="0" width="100" height="58" /></a></p>
</div>
</div>',
'label1' => '',
'info1' => '',
'label2' => 'Characteristics',
'info2' => '<ul>
<li><strong>Revolutionary:</strong> Only 10.000 cells needed for complete ChIP-seq procedure</li>
<li><strong>Reliable and accurate</strong> sequencing library preparation from just picogram inputs</li>
<li><strong>Validated on</strong> studies for histone marks and with the IP-Star<sup>®</sup> Compact Automated Platform</li>
</ul>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-histone-results.png" alt="ChIP on 10,000 cells" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>High efficiency ChIP on 10,000 cells</strong><br />ChIP efficiency on 10 000 cells. ChIP was performed on human Hela cells using the Diagenode antibodies H3K4me3 (Cat. No. pAb-003-050), H3K27ac (pAb-174-050), H3K9me3 (pAb-056-050) and H3K27me3 (pAb-069-050). Sheared chromatin from 10 000 cells and 0.1 µg (H3K27ac), 0.25 µg (H3K4me3 and H3K27me3) or 0.5 µg (H3K9me3) of the antibody were used per IP. Corresponding amount of IgG was used as control. Quantitative PCR was performed with primers for corresponding positive and negative loci. Figure shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-A.png" alt="ChIP amplification" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-B.png" alt="Match between True MicroChIP peaks and the reference dataset" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Reliable detection of enrichments in ChIP-seq</strong><br />A: ChIP has been peformed with H3K4me3 antibody, amplification of 17 pg of DNA ChIP'd from 10.000 cells and amplification of 35 pg of DNA ChIP'd from 100.000 cells (control experiment). The IP'd DNA was amplified and transformed into a sequencing-ready preparation for the Illumina plateform with the MicroPlex Library Preparation kit. The library was then analysed on an Illumina<sup>®</sup> Genome Analyzer. Cluster generation and sequencing were performed according to the manufacturer's instructions.<br />B: We observed a perfect match between the top 40% of True MicroChIP peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-result.png" alt="Cell lysis" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Hela cells were fixed with 1% formaldehyde (for 10 minutes at RT)</strong><br /> Cell lysis was performed using the Lysis Buffer tL1 of the Diagenode True MicroChIP kit. Samples corresponding to 10 000 cells are sheared during 5 rounds of 5 cycles of 30 seconds “ON” / 30 seconds “OFF” with the Bioruptor<sup>®</sup> Plus combined with the Bioruptor<sup>®</sup> Water cooler (Cat No. BioAcc-cool) at HIGH power setting (position H). For optimal results, samples are vortexed before and after performing 5 sonication cycles, followed by a short centrifugation at 4°C. 10 μl of DNA (equivalent to 60 000 cells) are analysed on a 1.5% agarose gel.</p>',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C01010140',
'old_catalog_number' => '',
'sf_code' => 'C01010140-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '540',
'price_USD' => '590',
'price_GBP' => '495',
'price_JPY' => '84590',
'price_CNY' => '',
'price_AUD' => '1475',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-true-microchip-kit-16-rxns',
'meta_title' => 'Auto True MicroChIP kit',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'modified' => '2019-05-10 09:28:26',
'created' => '2015-06-29 14:08:20',
'locale' => 'zho'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(),
'Group' => array(),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array()
)
)
$carrousel_order = array(
(int) 0 => 'ChIPmentation'
)
$language = 'cn'
$meta_keywords = ''
$meta_description = 'Auto True MicroChIP kit'
$meta_title = 'Auto True MicroChIP kit'
$product = array(
'Product' => array(
'id' => '1858',
'antibody_id' => null,
'name' => 'Auto True MicroChIP kit',
'description' => '<p>The Auto True MicroChIP kit in combination with the <a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns">MicroPlex Library Preparation™ kit</a> allows for performing ChIP-seq on as few as 10,000 cells. This microChIP-seq assay has been validated with the new generation <a href="../p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star<sup>®</sup> Compact Automated Workstation</a>. Furthermore Diagenode's high quality ChIP-seq grade antibodies have been used.</p>
<div class="row">
<div class="small-12 medium-5 large-6 columns"><small style="color: #666;">Video article</small>
<div class="flex-video"><iframe width="420" height="315" style="float: left; margin-right: 20px;" src="//www.youtube.com/embed/JZC9ARbLh_U?rel=0" frameborder="0" allowfullscreen="allowfullscreen"></iframe></div>
</div>
<div class="small-12 medium-7 large-6 columns">
<h3>Automating ChIP-seq Experiments to Generate Epigenetic Profiles on 10,000 HeLa Cells</h3>
<p><a href="https://www.youtube.com/watch?v=JZC9ARbLh_U"><img src="https://www.diagenode.com/img/jove_logo.png" alt="Jove logo" border="0" width="100" height="58" /></a></p>
</div>
</div>',
'label1' => '',
'info1' => '',
'label2' => 'Characteristics',
'info2' => '<ul>
<li><strong>Revolutionary:</strong> Only 10.000 cells needed for complete ChIP-seq procedure</li>
<li><strong>Reliable and accurate</strong> sequencing library preparation from just picogram inputs</li>
<li><strong>Validated on</strong> studies for histone marks and with the IP-Star<sup>®</sup> Compact Automated Platform</li>
</ul>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-histone-results.png" alt="ChIP on 10,000 cells" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>High efficiency ChIP on 10,000 cells</strong><br />ChIP efficiency on 10 000 cells. ChIP was performed on human Hela cells using the Diagenode antibodies H3K4me3 (Cat. No. pAb-003-050), H3K27ac (pAb-174-050), H3K9me3 (pAb-056-050) and H3K27me3 (pAb-069-050). Sheared chromatin from 10 000 cells and 0.1 µg (H3K27ac), 0.25 µg (H3K4me3 and H3K27me3) or 0.5 µg (H3K9me3) of the antibody were used per IP. Corresponding amount of IgG was used as control. Quantitative PCR was performed with primers for corresponding positive and negative loci. Figure shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-A.png" alt="ChIP amplification" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-B.png" alt="Match between True MicroChIP peaks and the reference dataset" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Reliable detection of enrichments in ChIP-seq</strong><br />A: ChIP has been peformed with H3K4me3 antibody, amplification of 17 pg of DNA ChIP'd from 10.000 cells and amplification of 35 pg of DNA ChIP'd from 100.000 cells (control experiment). The IP'd DNA was amplified and transformed into a sequencing-ready preparation for the Illumina plateform with the MicroPlex Library Preparation kit. The library was then analysed on an Illumina<sup>®</sup> Genome Analyzer. Cluster generation and sequencing were performed according to the manufacturer's instructions.<br />B: We observed a perfect match between the top 40% of True MicroChIP peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-result.png" alt="Cell lysis" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Hela cells were fixed with 1% formaldehyde (for 10 minutes at RT)</strong><br /> Cell lysis was performed using the Lysis Buffer tL1 of the Diagenode True MicroChIP kit. Samples corresponding to 10 000 cells are sheared during 5 rounds of 5 cycles of 30 seconds “ON” / 30 seconds “OFF” with the Bioruptor<sup>®</sup> Plus combined with the Bioruptor<sup>®</sup> Water cooler (Cat No. BioAcc-cool) at HIGH power setting (position H). For optimal results, samples are vortexed before and after performing 5 sonication cycles, followed by a short centrifugation at 4°C. 10 μl of DNA (equivalent to 60 000 cells) are analysed on a 1.5% agarose gel.</p>',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C01010140',
'old_catalog_number' => '',
'sf_code' => 'C01010140-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '540',
'price_USD' => '590',
'price_GBP' => '495',
'price_JPY' => '84590',
'price_CNY' => '',
'price_AUD' => '1475',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-true-microchip-kit-16-rxns',
'meta_title' => 'Auto True MicroChIP kit',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'modified' => '2019-05-10 09:28:26',
'created' => '2015-06-29 14:08:20',
'locale' => 'zho'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(),
'Group' => array(),
'Related' => array(
(int) 0 => array(
'id' => '1885',
'antibody_id' => null,
'name' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'description' => '<p><span>The Auro hMeDIP kit is designed for enrichment of hydroxymethylated DNA from fragmented genomic DNA samples for use in genome-wide methylation analysis. It features</span><span> a highly specific monoclonal antibody against </span><span>5-hydroxymethylcytosine (5-hmC) for the immunoprecipitation of hydroxymethylated DNA</span><span>. It includes control DNA and primers to assess the effiency of the assay. </span><span>Performing hydroxymethylation profiling with the hMeDIP kit is fast, reliable and highly specific.</span></p>',
'label1' => ' Characteristics',
'info1' => '<ul style="list-style-type: disc;">
<li><span>Robust enrichment & immunoprecipitation of hydroxymethylated DNA</span></li>
<li>Highly specific monoclonal antibody against 5-hmC<span> for reliable, reproducible results</span></li>
<li>Including control DNA and primers to <span>monitor the efficiency of the assay</span>
<ul style="list-style-type: circle;">
<li>5-hmC, 5-mC and unmethylated DNA sequences and primer pairs</li>
<li>Mouse primer pairs against Sfi1 targeting hydroxymethylated gene in mouse</li>
</ul>
</li>
</ul>
<ul style="list-style-type: disc;">
<li>Improved single-tube, magnetic bead-based protocol</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C02010034',
'old_catalog_number' => '',
'sf_code' => 'C02010034-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '630',
'price_USD' => '690',
'price_GBP' => '580',
'price_JPY' => '98690',
'price_CNY' => '',
'price_AUD' => '1725',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns',
'meta_title' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'meta_keywords' => '',
'meta_description' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'modified' => '2021-01-18 10:37:19',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1893',
'antibody_id' => null,
'name' => 'Auto Premium Bisulfite kit',
'description' => '<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => 'Characteristics',
'info1' => '<p><strong>Bisulfite Conversion efficiency in TERTBS1 and CTS56 genomic regions.</strong></p>
<p>The figure shows the conversion efficiency in TERTBS1 and CTS56 genomic regions when using 1 μg, 500ng and 100 ng of genomic DNA. Between 1 and 5 clones were analyzed for the different amounts of genomic DNA</p>
<p><img src="https://www.diagenode.com/img/product/kits/auto-premium-bisulfite-clone.png" alt="Clone" /></p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '40 rxns',
'catalog_number' => 'C02030031',
'old_catalog_number' => '',
'sf_code' => 'C02030031-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-premium-bisulfite-kit-40-rxns',
'meta_title' => 'Auto Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Auto Premium Bisulfite kit',
'modified' => '2019-12-16 10:05:26',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1842',
'antibody_id' => null,
'name' => 'Auto iDeal ChIP-seq Kit for Transcription Factors',
'description' => '<p><span><strong>This product must be used with the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star Compact Automated System</a>.</strong></span></p>
<p><span>Diagenode’s </span><strong>Auto iDeal ChIP-seq Kit for Transcription Factors</strong><span> is a highly specialized solution for robust Transcription Factor ChIP-seq results. Unlike competing solutions, our kit utilizes a highly optimized protocol and is backed by validation with a broad number and range of transcription factors. The kit provides high yields with excellent specificity and sensitivity.</span></p>',
'label1' => 'Characteristics',
'info1' => '<ul>
<li><strong>Confidence in results:</strong> Validated for ChIP-seq with multiple transcription factors</li>
<li><strong>Proven:</strong> Validated by the epigenetics community, including the BLUEPRINT consortium</li>
<li><strong>Most complete kit available</strong> for highest quality data - includes control antibodies and primers</li>
<li>Validated with Diagenode's <a href="https://www.diagenode.com/en/p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns"><span>MicroPlex Library Preparation™ kit</span></a> and <a href="https://www.diagenode.com/categories/ip-star" title="IP-Star Automated System">IP-Star<sup>®</sup></a> Automation System</li>
</ul>
<p> </p>
<h3>ChIP-seq on cells</h3>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-ctcf-diagenode.jpg" alt="CTCF Diagenode" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1.</strong> (A) Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina<sup>®</sup> HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the GAPDH positive control gene.</p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-figure-b-total-diagendoe-peaks.png" alt="CTCF Diagenode" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1B.</strong> The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p> </p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-A.png" alt="ChIP-seq figure A" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-B.png" alt="ChIP-seq figure B" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-C.png" alt="ChIP-seq figure C" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 2.</strong> Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade HDAC1 (A), LSD1 (B) and p53 antibody (C). The IP'd DNA was subsequently analysed on an Illumina<sup>®</sup> Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in regions of chromosome 3 (A), chromosome 12 (B) and chromosome 6 (C) respectively.</p>
<p> </p>
<h3>ChIP-seq on tissue</h3>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-figure-3a.jpg" alt="ChIP-seq figure A" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 3A.</strong> Chromatin Immunoprecipitation has been performed using chromatin from mouse liver tissue, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina® HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the Vwf positive control gene.</p>
<p><img src="https://www.diagenode.com/img/product/kits/match-of-the-top40-peaks.png" alt="Match of the Top40 peaks" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 3B.</strong> The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>',
'label2' => 'Species, cell lines, tissues tested',
'info2' => '<p>The iDeal ChIP-seq Kit for Transcription Factors is compatible with a broad variety of cell lines, tissues and species, as shown below. Other species / cell lines / tissues can be used with this kit.</p>
<p><span style="text-decoration: underline;">Cell lines:</span></p>
<p>Human: A549, A673, BT-549, CD4 T, HCC1806, HeLa, HepG2, HFF, HK-GFP-MR, ILC, K562, KYSE-180, LapC4, M14, MCF7, MDA-MB-231, MDA-MB-436, RDES, SKNO1, VCaP, U2-OS, ZR-75-1 </p>
<p>Mouse: ESC, NPCs, BZ, GT1-7, acinar cells, HSPCs, Th2 cells, keratinocytes</p>
<p>Cattle: pbMEC, <span>MAC-T</span></p>
<p><span style="text-decoration: underline;">Tissues:</span></p>
<p>Mouse: kidney, heart, brain, iris, liver, limbs from E10.5 embryos</p>
<p><span>Horse: l</span>iver, brain, heart, lung, skeletal muscle, lamina, ovary</p>
<p><span style="text-decoration: underline;">ChIP on yeast</span></p>
<p>The iDeal ChIP-seq kit for TF is compatible with yeast samples. Check out our <strong><a href="https://www.diagenode.com/files/products/kits/Application_Note-ChIP_on_Yeast.pdf">Application Note</a></strong> presenting an optimized detailed protocol for ChIP on yeast.</p>
<p></p>
<p>Did you use the iDeal ChIP-seq for Transcription Factors Kit on other cell line / tissue / species? <a href="mailto:agnieszka.zelisko@diagenode.com?subject=Species, cell lines, tissues tested with the iDeal ChIP-seq Kit for TF&body=Dear Customer,%0D%0A%0D%0APlease, leave below your feedback about the iDeal ChIP-seq for Transcription Factors (cell / tissue type, species, other information...).%0D%0A%0D%0AThank you for sharing with us your experience !%0D%0A%0D%0ABest regards,%0D%0A%0D%0AAgnieszka Zelisko-Schmidt, PhD">Let us know!</a></p>',
'label3' => 'Additional solutions compatible with Auto iDeal ChIP-seq kit for Transcription Factors',
'info3' => '<p><span style="font-weight: 400;">The</span> <a href="https://www.diagenode.com/en/p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns"><span style="font-weight: 400;">Chromatin shearing optimization kit – Low SDS (iDeal Kit for TFs)</span></a><span style="font-weight: 400;"> is the kit compatible with the iDeal ChIP-seq kit for TF, recommended for the optimization of chromatin shearing, a critical step for ChIP.</span></p>
<p><a href="https://www.diagenode.com/en/p/chip-cross-link-gold-600-ul"><span style="font-weight: 400;">ChIP Cross-link Gold</span></a> <span style="font-weight: 400;">should be used in combination with formaldehyde when working with higher order and/or dynamic interactions, for efficient protein-protein fixation.</span></p>
<p><span style="font-weight: 400;">For library preparation of immunoprecipitated samples we recommend to use the </span><b> </b><a href="https://www.diagenode.com/en/categories/library-preparation-for-ChIP-seq"><span style="font-weight: 400;">MicroPlex Library Preparation Kit</span></a><span style="font-weight: 400;"> - validated for library preparation from picogram inputs.</span></p>
<p><a href="https://www.diagenode.com/en/categories/chip-seq-grade-antibodies"><span style="font-weight: 400;">ChIP-seq grade antibodies</span></a><span style="font-weight: 400;"> provide high yields with excellent specificity and sensitivity.</span></p>
<p><span style="font-weight: 400;">Check the list of available </span><a href="https://www.diagenode.com/en/categories/primer-pairs"><span style="font-weight: 400;">Primer pairs</span></a><span style="font-weight: 400;"> designed for high specificity to specific genomic regions.</span></p>',
'format' => '24 rxns',
'catalog_number' => 'C01010058',
'old_catalog_number' => '',
'sf_code' => 'C01010058-',
'type' => 'RFR',
'search_order' => '01-Accessory',
'price_EUR' => '915',
'price_USD' => '1130',
'price_GBP' => '840',
'price_JPY' => '143335',
'price_CNY' => '',
'price_AUD' => '2825',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns',
'meta_title' => 'Auto iDeal ChIP-seq Kit for Transcription Factors x24',
'meta_keywords' => '',
'meta_description' => 'Auto iDeal ChIP-seq Kit for Transcription Factors x24',
'modified' => '2021-11-23 10:51:46',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1923',
'antibody_id' => null,
'name' => 'MicroChIP DiaPure columns',
'description' => '<p><a href="https://www.diagenode.com/files/products/reagents/MicroChIP_DiaPure_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p>Diagenode’s MicroChIP DiaPure columns have been optimized for the purification and elution of very low amounts of DNA. This rapid method has been validated for epigenetic applications like low input ChIP (e.g. using the True MicroChIP kit) and CUT&Tag (e.g. using Diagenode’s pA-Tn5), but is also compatible with many other applications. The DNA can be eluted at high concentrations in volumes down to 6 μl and it is suitable for any downstream application (e.g. NGS).</p>
<p>Benefits of the MicroChIP DiaPure columns:</p>
<ul>
<li>Optimized for the purification of very low DNA amounts</li>
<li>Fast and easy protocol</li>
<li>Non-toxic</li>
<li>Validated for ChIP and Cut&Tag</li>
</ul>',
'label1' => 'Examples of results',
'info1' => '<h2 style="text-align: center;">MicroChIP DiaPure columns after ChIP</h2>
<p>Successful ChIP-seq results generated on 50,000 of K562 cells using True MicroChIP technology. ChIP has been performed accordingly to True MicroChIP protocol (Diagenode, Cat. No. C01010130), including DNA purification using the MicroChIP DiaPure columns. For the library preparation the MicroPlex Library Preparation Kit (Diagenode, Cat. No. C05010001) has been used. The below figure shows the peaks from ChIP-seq experiments using the following Diagenode antibodies: H3K4me1 (C15410194), H3K9/14ac (C15410200), H3K27ac (C15410196) and H3K36me3 (C15410192).</p>
<p><img src="https://www.diagenode.com/img/product/kits/figure-igv-microchip.png" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1:</strong> Integrative genomics viewer (IGV) visualization of ChIP-seq experiments using 50,000 of K562 cells.</p>
<p></p>
<h2 style="text-align: center;"></h2>
<h2 style="text-align: center;">MicroChIP DiaPure columns after CUT&Tag</h2>
<p>Successful CUT&Tag results showing a low background with high region-specific enrichment has been generated using 50.000 of K562 cells, 1 µg of H3K27me3 antibody (Diagenode, Cat. No. C15410069) and proteinA-Tn5 (1:250) (Diagenode, Cat. No. C01070001). 1 µg of IgG (Diagenode, Cat. No. C15410206) was used as negative control. Samples were purified using the MicroChIP DiaPure columns or phenol-chloroform purification. The below figure presenst the comparison of two purification methods.</p>
<p><img src="https://www.diagenode.com/img/product/kits/figure-diapure-igv.png" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 2:</strong> Integrative genomics viewer (IGV) visualization of CUT&Tag experiments: MicroChIP DiaPure columns vs phenol-chloroform purification using the H3K27me3 antibody.</p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C03040001',
'old_catalog_number' => '',
'sf_code' => 'C03040001-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'microchip-diapure-columns-50-rxns',
'meta_title' => 'MicroChIP DiaPure columns',
'meta_keywords' => '',
'meta_description' => 'MicroChIP DiaPure columns',
'modified' => '2023-04-20 16:11:26',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array([maximum depth reached])
),
(int) 4 => array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '9',
'position' => '10',
'parent_id' => '2',
'name' => 'ChIP-seq',
'description' => '<div class="row">
<div class="large-12 columns">Chromatin Immunoprecipitation (ChIP) coupled with high-throughput massively parallel sequencing as a detection method (ChIP-seq) has become one of the primary methods for epigenomics researchers, namely to investigate protein-DNA interaction on a genome-wide scale. This technique is now used in a variety of life science disciplines including cellular differentiation, tumor suppressor gene silencing, and the effect of histone modifications on gene expression.</div>
<div class="large-12 columns"></div>
<h5 class="large-12 columns"><strong></strong></h5>
<h5 class="large-12 columns"><strong>The ChIP-seq workflow</strong></h5>
<div class="small-12 medium-12 large-12 columns text-center"><br /><img src="https://www.diagenode.com/img/chip-seq-diagram.png" /></div>
<div class="large-12 columns"><br />
<ol>
<li class="large-12 columns"><strong>Chromatin preparation: </strong>Crosslink chromatin-bound proteins (histones or transcription factors) to DNA followed by cell lysis.</li>
<li class="large-12 columns"><strong>Chromatin shearing:</strong> Fragment chromatin by sonication to desired fragment size (100-500 bp)</li>
<li class="large-12 columns"><strong>Chromatin IP</strong>: Capture protein-DNA complexes with <strong><a href="../categories/chip-seq-grade-antibodies">specific ChIP-seq grade antibodies</a></strong> against the histone or transcription factor of interest</li>
<li class="large-12 columns"><strong>DNA purification</strong>: Reverse cross-links, elute, and purify </li>
<li class="large-12 columns"><strong>NGS Library Preparation</strong>: Ligate adapters and amplify IP'd material</li>
<li class="large-12 columns"><strong>Bioinformatic analysis</strong>: Perform r<span style="font-weight: 400;">ead filtering and trimming</span>, r<span style="font-weight: 400;">ead specific alignment, enrichment specific peak calling, QC metrics, multi-sample cross-comparison etc. </span></li>
</ol>
</div>
</div>
<div class="row" style="margin-top: 32px;">
<div class="small-12 medium-10 large-9 small-centered columns">
<div class="radius panel" style="background-color: #fff;">
<h3 class="text-center" style="color: #b21329;">Need guidance?</h3>
<p class="text-justify">Choose our full ChIP kits or simply choose what you need from antibodies, buffers, beads, chromatin shearing and purification reagents. With the ChIP Kit Customizer, you have complete flexibility on which components you want from our validated ChIP kits.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns"><a href="../pages/which-kit-to-choose"><img alt="" src="https://www.diagenode.com/img/banners/banner-decide.png" /></a></div>
<div class="small-6 medium-6 large-6 columns"><a href="../pages/chip-kit-customizer-1"><img alt="" src="https://www.diagenode.com/img/banners/banner-customizer.png" /></a></div>
</div>
</div>
</div>
</div>',
'in_footer' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'slug' => 'chromatin-immunoprecipitation-sequencing',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers wide range of kits and antibodies for Chromatin Immunoprecipitation Sequencing (ChIP-Seq) and also provides Bioruptor for chromatin shearing',
'meta_title' => 'Chromatin Immunoprecipitation - ChIP-seq Kits - Dna methylation | Diagenode',
'modified' => '2017-11-14 09:57:16',
'created' => '2015-04-12 18:08:46',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '117',
'position' => '3',
'parent_id' => '59',
'name' => 'Automated kits',
'description' => '',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'hide' => false,
'all_format' => false,
'is_antibody' => false,
'slug' => 'chromatin-ip-automated-kits',
'cookies_tag_id' => null,
'meta_keywords' => '',
'meta_description' => 'Diagenode’s validated ChIP-Seq Automated kits has everything you need for a successful ChIP prior to Next-Generation Sequencing.',
'meta_title' => 'Automated ChIP-Seq Kits for Chromatin Immunoprecipitation | Diagenode',
'modified' => '2022-02-11 14:11:01',
'created' => '2016-07-19 16:57:20',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '110',
'name' => 'Auto TrueMicroChIP kit',
'description' => '<div class="page" title="Page 4">
<div class="layoutArea">
<div class="column">
<p><span></span><span>Conventional ChIP protocols require high numbers of cells (hundreds of thousands cells at least) limiting the application for ChIP technology to few cell samples. Recently, ChIP assays on smallest amount of cells have been reported. The procedure requires tedious optimization of several reaction conditions to face the increased background observed in ChIP performed with reduced amount of cells. That might consequently lead to considerable time and lab expenditures. To reduce these tedious steps, Diagenode provides the new Auto True MicroChIP kit with optimized reagents and protocol to enable successful ChIP on as few as 10 000 cells. Moreover, the Auto True MicroChIP kit protocol has been thoroughly optimised by Diagenode for ChIP followed by high-throughput sequencing on Illumina</span><span>® </span><span>Next-Gen sequencers. To allow the generation of consistent results in ChIP-seq on 10 000 cells, Diagenode has also optimised a library preparation protocol on limited amount of immunoprecipitated DNA. </span></p>
<p><span>The new MicroPLEX library preparation kit allows the preparation of libraries for sequencing on picogram amount of immunoprecipitated DNA. Thus, association of the Auto True MicroChIP kit with the MicroPLEX library preparation kit provides optimised solutions to perform ChIP-sequencing on limited amount of cells. </span></p>
</div>
</div>
</div>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/Auto-TrueMicroChIP-kit-manual.pdf',
'slug' => 'auto-truemicrochip-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-09-01 12:30:58',
'created' => '2015-07-07 11:47:43',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '37',
'name' => 'Chromatin Brochure',
'description' => '<p>Whether you are experienced or new to the field of chromatin immunoprecipitation, Diagenode has everything you need to make ChIP easy and convenient while ensuring consistent data between samples and experiments. As an expert in the field of epigenetics, Diagenode is committed to providing complete solutions from chromatin shearing reagents, shearing instruments such as the Bioruptor® (the gold standard for chromatin shearing), ChIP kits, the largest number of validated and trusted antibodies on the market, and the SX-8G IP-Star® Compact Automated System to achieve unparalleled productivity and reproducibility.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf',
'slug' => 'chromatin-immunoprecipitation-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2022-03-24 12:34:11',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1775',
'name' => 'product/kits/chip-kit-icon.png',
'alt' => 'ChIP kit icon',
'modified' => '2018-04-17 11:52:29',
'created' => '2018-03-15 15:50:34',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '3563',
'name' => 'EZH2 is overexpressed in transitional preplasmablasts and is involved in human plasma cell differentiation.',
'authors' => 'Herviou L, Jourdan M, Martinez AM, Cavalli G, Moreaux J',
'description' => '<p>Plasma cells (PCs) play a major role in the defense of the host organism against pathogens. We have shown that PC generation can be modeled using multi-step culture systems that reproduce the sequential cell differentiation occurring in vivo. Using this unique model, we investigated the role of EZH2 during PC differentiation (PCD) using H3K27me3 and EZH2 ChIP-binding profiles. We then studied the effect of the inhibition of EZH2 enzymatic activity to understand how EZH2 regulates the key functions involved in PCD. EZH2 expression significantly increases in preplasmablasts with H3K27me3 mediated repression of genes involved in B cell and plasma cell identity. EZH2 was also found to be recruited to H3K27me3-free promoters of transcriptionally active genes known to regulate cell proliferation. Inhibition the catalytic activity of EZH2 resulted in B to PC transcriptional changes associated with PC maturation induction, as well as higher immunoglobulin secretion. Altogether, our data suggest that EZH2 is involved in the maintenance of preplasmablast transitory immature proliferative state that supports their amplification.</p>',
'date' => '2019-02-12',
'pmid' => 'http://www.pubmed.gov/30755708',
'doi' => '10.1038/s41375-019-0392-1',
'modified' => '2019-03-21 17:17:48',
'created' => '2019-03-21 14:12:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '3389',
'name' => 'An Integrated and Semiautomated Microscaled Approach to Profile Cis-Regulatory Elements by Histone Modification ChIP-Seq for Large-Scale Epigenetic Studies.',
'authors' => 'Youhanna Jankeel D, Cayford J, Schmiedel BJ, Vijayanand P, Seumois G.',
'description' => '<p>Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) is the preferred approach to map histone modifications and identify cis-regulatory DNA elements throughout the genome. Multiple methods have been described to increase the efficiency of library preparation and to reduce hands-on time as well as costs. This review describes detailed steps to perform cell fixation, chromatin shearing, immunoprecipitation, and sequencing library preparation for a batch of 48-96 samples with small cell numbers. The protocol implements a semiautomated platform to reduce technical variability and improve signal-to-noise ratio as well as reduce hands-on time, thus allowing large-scale epigenetic studies of clinical samples with limited cell numbers.</p>',
'date' => '2018-06-30',
'pmid' => 'http://www.pubmed.gov/29956160',
'doi' => '10.1007/978-1-4939-7896-0_22',
'modified' => '2018-11-09 12:29:33',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '3595',
'name' => 'Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche.',
'authors' => 'Baghdadi MB, Castel D, Machado L, Fukada SI, Birk DE, Relaix F, Tajbakhsh S, Mourikis P',
'description' => '<p>The cell microenvironment, which is critical for stem cell maintenance, contains both cellular and non-cellular components, including secreted growth factors and the extracellular matrix. Although Notch and other signalling pathways have previously been reported to regulate quiescence of stem cells, the composition and source of molecules that maintain the stem cell niche remain largely unknown. Here we show that adult muscle satellite (stem) cells in mice produce extracellular matrix collagens to maintain quiescence in a cell-autonomous manner. Using chromatin immunoprecipitation followed by sequencing, we identified NOTCH1/RBPJ-bound regulatory elements adjacent to specific collagen genes, the expression of which is deregulated in Notch-mutant mice. Moreover, we show that Collagen V (COLV) produced by satellite cells is a critical component of the quiescent niche, as depletion of COLV by conditional deletion of the Col5a1 gene leads to anomalous cell cycle entry and gradual diminution of the stem cell pool. Notably, the interaction of COLV with satellite cells is mediated by the Calcitonin receptor, for which COLV acts as a surrogate local ligand. Systemic administration of a calcitonin derivative is sufficient to rescue the quiescence and self-renewal defects found in COLV-null satellite cells. This study reveals a Notch-COLV-Calcitonin receptor signalling cascade that maintains satellite cells in a quiescent state in a cell-autonomous fashion, and raises the possibility that similar reciprocal mechanisms act in diverse stem cell populations.</p>',
'date' => '2018-05-23',
'pmid' => 'http://www.pubmed.gov/29795344',
'doi' => '10.1038/s41586-018-0144-9',
'modified' => '2019-04-17 15:12:55',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '3589',
'name' => 'A new metabolic gene signature in prostate cancer regulated by JMJD3 and EZH2.',
'authors' => 'Daures M, Idrissou M, Judes G, Rifaï K, Penault-Llorca F, Bignon YJ, Guy L, Bernard-Gallon D',
'description' => '<p>Histone methylation is essential for gene expression control. Trimethylated lysine 27 of histone 3 (H3K27me3) is controlled by the balance between the activities of JMJD3 demethylase and EZH2 methyltransferase. This epigenetic mark has been shown to be deregulated in prostate cancer, and evidence shows H3K27me3 enrichment on gene promoters in prostate cancer. To study the impact of this enrichment, a transcriptomic analysis with TaqMan Low Density Array (TLDA) of several genes was studied on prostate biopsies divided into three clinical grades: normal ( = 23) and two tumor groups that differed in their aggressiveness (Gleason score ≤ 7 ( = 20) and >7 ( = 19)). ANOVA demonstrated that expression of the gene set was upregulated in tumors and correlated with Gleason score, thus discriminating between the three clinical groups. Six genes involved in key cellular processes stood out: , , , , and . Chromatin immunoprecipitation demonstrated collocation of EZH2 and JMJD3 on gene promoters that was dependent on disease stage. Gene set expression was also evaluated on prostate cancer cell lines (DU 145, PC-3 and LNCaP) treated with an inhibitor of JMJD3 (GSK-J4) or EZH2 (DZNeP) to study their involvement in gene regulation. Results showed a difference in GSK-J4 sensitivity under PTEN status of cell lines and an opposite gene expression profile according to androgen status of cells. In summary, our data describe the impacts of JMJD3 and EZH2 on a new gene signature involved in prostate cancer that may help identify diagnostic and therapeutic targets in prostate cancer.</p>',
'date' => '2018-05-04',
'pmid' => 'http://www.pubmed.gov/29805743',
'doi' => '10.18632/oncotarget.25182',
'modified' => '2019-04-17 15:21:33',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '3193',
'name' => 'Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression',
'authors' => 'Ngollo M. et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">H3K27me3 histone marks shape the inhibition of gene transcription. In prostate cancer, the deregulation of H3K27me3 marks might play a role in prostate tumor progression.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">We investigated genome-wide H3K27me3 histone methylation profile using chromatin immunoprecipitation (ChIP) and 2X400K promoter microarrays to identify differentially-enriched regions in biopsy samples from prostate cancer patients. H3K27me3 marks were assessed in 34 prostate tumors: 11 with Gleason score > 7 (GS > 7), 10 with Gleason score ≤ 7 (GS ≤ 7), and 13 morphologically normal prostate samples.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">Here, H3K27me3 profiling identified an average of 386 enriched-genes on promoter regions in healthy control group versus 545 genes in GS ≤ 7 and 748 genes in GS > 7 group. We then ran a factorial discriminant analysis (FDA) and compared the enriched genes in prostate-tumor biopsies and normal biopsies using ANOVA to identify significantly differentially-enriched genes. The analysis identified ALG5, EXOSC8, CBX1, GRID2, GRIN3B, ING3, MYO1D, NPHP3-AS1, MSH6, FBXO11, SND1, SPATS2, TENM4 and TRA2A genes. These genes are possibly associated with prostate cancer. Notably, the H3K27me3 histone mark emerged as a novel regulatory mechanism in poor-prognosis prostate cancer.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Our findings point to epigenetic mark H3K27me3 as an important event in prostate carcinogenesis and progression. The results reported here provide new molecular insights into the pathogenesis of prostate cancer.</abstracttext></p>
</div>',
'date' => '2017-04-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28403887',
'doi' => '',
'modified' => '2017-06-19 14:07:35',
'created' => '2017-06-19 14:05:03',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array()
)
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array()
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010034</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1885" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1885" id="CartAdd/1885Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1885" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto hMeDIP kit x16 (monoclonal mouse antibody)</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns" data-reveal-id="cartModal-1885" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto hMeDIP kit x16 (monoclonal mouse antibody)</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-premium-bisulfite-kit-40-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02030031</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1893" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1893" id="CartAdd/1893Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1893" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto Premium Bisulfite kit</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto Premium Bisulfite kit',
'C02030031',
'240',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto Premium Bisulfite kit',
'C02030031',
'240',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-premium-bisulfite-kit-40-rxns" data-reveal-id="cartModal-1893" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto Premium Bisulfite kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns"><img src="/img/product/kits/chip-kit-icon.png" alt="ChIP kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C01010058</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1842" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1842" id="CartAdd/1842Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1842" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto iDeal ChIP-seq Kit for Transcription Factors</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto iDeal ChIP-seq Kit for Transcription Factors',
'C01010058',
'1130',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto iDeal ChIP-seq Kit for Transcription Factors',
'C01010058',
'1130',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns" data-reveal-id="cartModal-1842" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto iDeal ChIP-seq Kit for Transcription Factors</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/microchip-diapure-columns-50-rxns"><img src="/img/grey-logo.jpg" alt="default alt" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C03040001</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1923" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1923" id="CartAdd/1923Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1923" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MicroChIP DiaPure columns</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MicroChIP DiaPure columns',
'C03040001',
'125',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MicroChIP DiaPure columns',
'C03040001',
'125',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="microchip-diapure-columns-50-rxns" data-reveal-id="cartModal-1923" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MicroChIP DiaPure columns</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020011</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1888" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1888" id="CartAdd/1888Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1888" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto MethylCap kit</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1888" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto MethylCap kit</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '3624',
'product_id' => '1858',
'related_id' => '1888'
),
'Image' => array(
(int) 0 => array(
'id' => '1778',
'name' => 'product/kits/methyl-kit-icon.png',
'alt' => 'Methylation kit icon',
'modified' => '2019-04-23 15:17:01',
'created' => '2018-03-15 15:52:12',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$document = array(
'id' => '37',
'name' => 'Chromatin Brochure',
'description' => '<p>Whether you are experienced or new to the field of chromatin immunoprecipitation, Diagenode has everything you need to make ChIP easy and convenient while ensuring consistent data between samples and experiments. As an expert in the field of epigenetics, Diagenode is committed to providing complete solutions from chromatin shearing reagents, shearing instruments such as the Bioruptor® (the gold standard for chromatin shearing), ChIP kits, the largest number of validated and trusted antibodies on the market, and the SX-8G IP-Star® Compact Automated System to achieve unparalleled productivity and reproducibility.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf',
'slug' => 'chromatin-immunoprecipitation-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2022-03-24 12:34:11',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '1094',
'product_id' => '1858',
'document_id' => '37'
)
)
$publication = array(
'id' => '3193',
'name' => 'Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression',
'authors' => 'Ngollo M. et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">H3K27me3 histone marks shape the inhibition of gene transcription. In prostate cancer, the deregulation of H3K27me3 marks might play a role in prostate tumor progression.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">We investigated genome-wide H3K27me3 histone methylation profile using chromatin immunoprecipitation (ChIP) and 2X400K promoter microarrays to identify differentially-enriched regions in biopsy samples from prostate cancer patients. H3K27me3 marks were assessed in 34 prostate tumors: 11 with Gleason score > 7 (GS > 7), 10 with Gleason score ≤ 7 (GS ≤ 7), and 13 morphologically normal prostate samples.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">Here, H3K27me3 profiling identified an average of 386 enriched-genes on promoter regions in healthy control group versus 545 genes in GS ≤ 7 and 748 genes in GS > 7 group. We then ran a factorial discriminant analysis (FDA) and compared the enriched genes in prostate-tumor biopsies and normal biopsies using ANOVA to identify significantly differentially-enriched genes. The analysis identified ALG5, EXOSC8, CBX1, GRID2, GRIN3B, ING3, MYO1D, NPHP3-AS1, MSH6, FBXO11, SND1, SPATS2, TENM4 and TRA2A genes. These genes are possibly associated with prostate cancer. Notably, the H3K27me3 histone mark emerged as a novel regulatory mechanism in poor-prognosis prostate cancer.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Our findings point to epigenetic mark H3K27me3 as an important event in prostate carcinogenesis and progression. The results reported here provide new molecular insights into the pathogenesis of prostate cancer.</abstracttext></p>
</div>',
'date' => '2017-04-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28403887',
'doi' => '',
'modified' => '2017-06-19 14:07:35',
'created' => '2017-06-19 14:05:03',
'ProductsPublication' => array(
'id' => '2138',
'product_id' => '1858',
'publication_id' => '3193'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/28403887" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: message [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'carrousel_order' => array(
(int) 0 => 'ChIPmentation'
),
'language' => 'cn',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'meta_title' => 'Auto True MicroChIP kit',
'product' => array(
'Product' => array(
'id' => '1858',
'antibody_id' => null,
'name' => 'Auto True MicroChIP kit',
'description' => '<p>The Auto True MicroChIP kit in combination with the <a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns">MicroPlex Library Preparation™ kit</a> allows for performing ChIP-seq on as few as 10,000 cells. This microChIP-seq assay has been validated with the new generation <a href="../p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star<sup>®</sup> Compact Automated Workstation</a>. Furthermore Diagenode's high quality ChIP-seq grade antibodies have been used.</p>
<div class="row">
<div class="small-12 medium-5 large-6 columns"><small style="color: #666;">Video article</small>
<div class="flex-video"><iframe width="420" height="315" style="float: left; margin-right: 20px;" src="//www.youtube.com/embed/JZC9ARbLh_U?rel=0" frameborder="0" allowfullscreen="allowfullscreen"></iframe></div>
</div>
<div class="small-12 medium-7 large-6 columns">
<h3>Automating ChIP-seq Experiments to Generate Epigenetic Profiles on 10,000 HeLa Cells</h3>
<p><a href="https://www.youtube.com/watch?v=JZC9ARbLh_U"><img src="https://www.diagenode.com/img/jove_logo.png" alt="Jove logo" border="0" width="100" height="58" /></a></p>
</div>
</div>',
'label1' => '',
'info1' => '',
'label2' => 'Characteristics',
'info2' => '<ul>
<li><strong>Revolutionary:</strong> Only 10.000 cells needed for complete ChIP-seq procedure</li>
<li><strong>Reliable and accurate</strong> sequencing library preparation from just picogram inputs</li>
<li><strong>Validated on</strong> studies for histone marks and with the IP-Star<sup>®</sup> Compact Automated Platform</li>
</ul>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-histone-results.png" alt="ChIP on 10,000 cells" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>High efficiency ChIP on 10,000 cells</strong><br />ChIP efficiency on 10 000 cells. ChIP was performed on human Hela cells using the Diagenode antibodies H3K4me3 (Cat. No. pAb-003-050), H3K27ac (pAb-174-050), H3K9me3 (pAb-056-050) and H3K27me3 (pAb-069-050). Sheared chromatin from 10 000 cells and 0.1 µg (H3K27ac), 0.25 µg (H3K4me3 and H3K27me3) or 0.5 µg (H3K9me3) of the antibody were used per IP. Corresponding amount of IgG was used as control. Quantitative PCR was performed with primers for corresponding positive and negative loci. Figure shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-A.png" alt="ChIP amplification" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-B.png" alt="Match between True MicroChIP peaks and the reference dataset" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Reliable detection of enrichments in ChIP-seq</strong><br />A: ChIP has been peformed with H3K4me3 antibody, amplification of 17 pg of DNA ChIP'd from 10.000 cells and amplification of 35 pg of DNA ChIP'd from 100.000 cells (control experiment). The IP'd DNA was amplified and transformed into a sequencing-ready preparation for the Illumina plateform with the MicroPlex Library Preparation kit. The library was then analysed on an Illumina<sup>®</sup> Genome Analyzer. Cluster generation and sequencing were performed according to the manufacturer's instructions.<br />B: We observed a perfect match between the top 40% of True MicroChIP peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-result.png" alt="Cell lysis" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Hela cells were fixed with 1% formaldehyde (for 10 minutes at RT)</strong><br /> Cell lysis was performed using the Lysis Buffer tL1 of the Diagenode True MicroChIP kit. Samples corresponding to 10 000 cells are sheared during 5 rounds of 5 cycles of 30 seconds “ON” / 30 seconds “OFF” with the Bioruptor<sup>®</sup> Plus combined with the Bioruptor<sup>®</sup> Water cooler (Cat No. BioAcc-cool) at HIGH power setting (position H). For optimal results, samples are vortexed before and after performing 5 sonication cycles, followed by a short centrifugation at 4°C. 10 μl of DNA (equivalent to 60 000 cells) are analysed on a 1.5% agarose gel.</p>',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C01010140',
'old_catalog_number' => '',
'sf_code' => 'C01010140-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '540',
'price_USD' => '590',
'price_GBP' => '495',
'price_JPY' => '84590',
'price_CNY' => '',
'price_AUD' => '1475',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-true-microchip-kit-16-rxns',
'meta_title' => 'Auto True MicroChIP kit',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'modified' => '2019-05-10 09:28:26',
'created' => '2015-06-29 14:08:20',
'locale' => 'zho'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(),
'Group' => array(),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array()
)
)
$carrousel_order = array(
(int) 0 => 'ChIPmentation'
)
$language = 'cn'
$meta_keywords = ''
$meta_description = 'Auto True MicroChIP kit'
$meta_title = 'Auto True MicroChIP kit'
$product = array(
'Product' => array(
'id' => '1858',
'antibody_id' => null,
'name' => 'Auto True MicroChIP kit',
'description' => '<p>The Auto True MicroChIP kit in combination with the <a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns">MicroPlex Library Preparation™ kit</a> allows for performing ChIP-seq on as few as 10,000 cells. This microChIP-seq assay has been validated with the new generation <a href="../p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star<sup>®</sup> Compact Automated Workstation</a>. Furthermore Diagenode's high quality ChIP-seq grade antibodies have been used.</p>
<div class="row">
<div class="small-12 medium-5 large-6 columns"><small style="color: #666;">Video article</small>
<div class="flex-video"><iframe width="420" height="315" style="float: left; margin-right: 20px;" src="//www.youtube.com/embed/JZC9ARbLh_U?rel=0" frameborder="0" allowfullscreen="allowfullscreen"></iframe></div>
</div>
<div class="small-12 medium-7 large-6 columns">
<h3>Automating ChIP-seq Experiments to Generate Epigenetic Profiles on 10,000 HeLa Cells</h3>
<p><a href="https://www.youtube.com/watch?v=JZC9ARbLh_U"><img src="https://www.diagenode.com/img/jove_logo.png" alt="Jove logo" border="0" width="100" height="58" /></a></p>
</div>
</div>',
'label1' => '',
'info1' => '',
'label2' => 'Characteristics',
'info2' => '<ul>
<li><strong>Revolutionary:</strong> Only 10.000 cells needed for complete ChIP-seq procedure</li>
<li><strong>Reliable and accurate</strong> sequencing library preparation from just picogram inputs</li>
<li><strong>Validated on</strong> studies for histone marks and with the IP-Star<sup>®</sup> Compact Automated Platform</li>
</ul>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-histone-results.png" alt="ChIP on 10,000 cells" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>High efficiency ChIP on 10,000 cells</strong><br />ChIP efficiency on 10 000 cells. ChIP was performed on human Hela cells using the Diagenode antibodies H3K4me3 (Cat. No. pAb-003-050), H3K27ac (pAb-174-050), H3K9me3 (pAb-056-050) and H3K27me3 (pAb-069-050). Sheared chromatin from 10 000 cells and 0.1 µg (H3K27ac), 0.25 µg (H3K4me3 and H3K27me3) or 0.5 µg (H3K9me3) of the antibody were used per IP. Corresponding amount of IgG was used as control. Quantitative PCR was performed with primers for corresponding positive and negative loci. Figure shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-A.png" alt="ChIP amplification" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-B.png" alt="Match between True MicroChIP peaks and the reference dataset" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Reliable detection of enrichments in ChIP-seq</strong><br />A: ChIP has been peformed with H3K4me3 antibody, amplification of 17 pg of DNA ChIP'd from 10.000 cells and amplification of 35 pg of DNA ChIP'd from 100.000 cells (control experiment). The IP'd DNA was amplified and transformed into a sequencing-ready preparation for the Illumina plateform with the MicroPlex Library Preparation kit. The library was then analysed on an Illumina<sup>®</sup> Genome Analyzer. Cluster generation and sequencing were performed according to the manufacturer's instructions.<br />B: We observed a perfect match between the top 40% of True MicroChIP peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-result.png" alt="Cell lysis" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Hela cells were fixed with 1% formaldehyde (for 10 minutes at RT)</strong><br /> Cell lysis was performed using the Lysis Buffer tL1 of the Diagenode True MicroChIP kit. Samples corresponding to 10 000 cells are sheared during 5 rounds of 5 cycles of 30 seconds “ON” / 30 seconds “OFF” with the Bioruptor<sup>®</sup> Plus combined with the Bioruptor<sup>®</sup> Water cooler (Cat No. BioAcc-cool) at HIGH power setting (position H). For optimal results, samples are vortexed before and after performing 5 sonication cycles, followed by a short centrifugation at 4°C. 10 μl of DNA (equivalent to 60 000 cells) are analysed on a 1.5% agarose gel.</p>',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C01010140',
'old_catalog_number' => '',
'sf_code' => 'C01010140-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '540',
'price_USD' => '590',
'price_GBP' => '495',
'price_JPY' => '84590',
'price_CNY' => '',
'price_AUD' => '1475',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-true-microchip-kit-16-rxns',
'meta_title' => 'Auto True MicroChIP kit',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'modified' => '2019-05-10 09:28:26',
'created' => '2015-06-29 14:08:20',
'locale' => 'zho'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(),
'Group' => array(),
'Related' => array(
(int) 0 => array(
'id' => '1885',
'antibody_id' => null,
'name' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'description' => '<p><span>The Auro hMeDIP kit is designed for enrichment of hydroxymethylated DNA from fragmented genomic DNA samples for use in genome-wide methylation analysis. It features</span><span> a highly specific monoclonal antibody against </span><span>5-hydroxymethylcytosine (5-hmC) for the immunoprecipitation of hydroxymethylated DNA</span><span>. It includes control DNA and primers to assess the effiency of the assay. </span><span>Performing hydroxymethylation profiling with the hMeDIP kit is fast, reliable and highly specific.</span></p>',
'label1' => ' Characteristics',
'info1' => '<ul style="list-style-type: disc;">
<li><span>Robust enrichment & immunoprecipitation of hydroxymethylated DNA</span></li>
<li>Highly specific monoclonal antibody against 5-hmC<span> for reliable, reproducible results</span></li>
<li>Including control DNA and primers to <span>monitor the efficiency of the assay</span>
<ul style="list-style-type: circle;">
<li>5-hmC, 5-mC and unmethylated DNA sequences and primer pairs</li>
<li>Mouse primer pairs against Sfi1 targeting hydroxymethylated gene in mouse</li>
</ul>
</li>
</ul>
<ul style="list-style-type: disc;">
<li>Improved single-tube, magnetic bead-based protocol</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C02010034',
'old_catalog_number' => '',
'sf_code' => 'C02010034-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '630',
'price_USD' => '690',
'price_GBP' => '580',
'price_JPY' => '98690',
'price_CNY' => '',
'price_AUD' => '1725',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns',
'meta_title' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'meta_keywords' => '',
'meta_description' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'modified' => '2021-01-18 10:37:19',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1893',
'antibody_id' => null,
'name' => 'Auto Premium Bisulfite kit',
'description' => '<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => 'Characteristics',
'info1' => '<p><strong>Bisulfite Conversion efficiency in TERTBS1 and CTS56 genomic regions.</strong></p>
<p>The figure shows the conversion efficiency in TERTBS1 and CTS56 genomic regions when using 1 μg, 500ng and 100 ng of genomic DNA. Between 1 and 5 clones were analyzed for the different amounts of genomic DNA</p>
<p><img src="https://www.diagenode.com/img/product/kits/auto-premium-bisulfite-clone.png" alt="Clone" /></p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '40 rxns',
'catalog_number' => 'C02030031',
'old_catalog_number' => '',
'sf_code' => 'C02030031-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-premium-bisulfite-kit-40-rxns',
'meta_title' => 'Auto Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Auto Premium Bisulfite kit',
'modified' => '2019-12-16 10:05:26',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1842',
'antibody_id' => null,
'name' => 'Auto iDeal ChIP-seq Kit for Transcription Factors',
'description' => '<p><span><strong>This product must be used with the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star Compact Automated System</a>.</strong></span></p>
<p><span>Diagenode’s </span><strong>Auto iDeal ChIP-seq Kit for Transcription Factors</strong><span> is a highly specialized solution for robust Transcription Factor ChIP-seq results. Unlike competing solutions, our kit utilizes a highly optimized protocol and is backed by validation with a broad number and range of transcription factors. The kit provides high yields with excellent specificity and sensitivity.</span></p>',
'label1' => 'Characteristics',
'info1' => '<ul>
<li><strong>Confidence in results:</strong> Validated for ChIP-seq with multiple transcription factors</li>
<li><strong>Proven:</strong> Validated by the epigenetics community, including the BLUEPRINT consortium</li>
<li><strong>Most complete kit available</strong> for highest quality data - includes control antibodies and primers</li>
<li>Validated with Diagenode's <a href="https://www.diagenode.com/en/p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns"><span>MicroPlex Library Preparation™ kit</span></a> and <a href="https://www.diagenode.com/categories/ip-star" title="IP-Star Automated System">IP-Star<sup>®</sup></a> Automation System</li>
</ul>
<p> </p>
<h3>ChIP-seq on cells</h3>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-ctcf-diagenode.jpg" alt="CTCF Diagenode" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1.</strong> (A) Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina<sup>®</sup> HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the GAPDH positive control gene.</p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-figure-b-total-diagendoe-peaks.png" alt="CTCF Diagenode" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1B.</strong> The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p> </p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-A.png" alt="ChIP-seq figure A" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-B.png" alt="ChIP-seq figure B" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-C.png" alt="ChIP-seq figure C" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 2.</strong> Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade HDAC1 (A), LSD1 (B) and p53 antibody (C). The IP'd DNA was subsequently analysed on an Illumina<sup>®</sup> Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in regions of chromosome 3 (A), chromosome 12 (B) and chromosome 6 (C) respectively.</p>
<p> </p>
<h3>ChIP-seq on tissue</h3>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-figure-3a.jpg" alt="ChIP-seq figure A" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 3A.</strong> Chromatin Immunoprecipitation has been performed using chromatin from mouse liver tissue, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina® HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the Vwf positive control gene.</p>
<p><img src="https://www.diagenode.com/img/product/kits/match-of-the-top40-peaks.png" alt="Match of the Top40 peaks" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 3B.</strong> The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>',
'label2' => 'Species, cell lines, tissues tested',
'info2' => '<p>The iDeal ChIP-seq Kit for Transcription Factors is compatible with a broad variety of cell lines, tissues and species, as shown below. Other species / cell lines / tissues can be used with this kit.</p>
<p><span style="text-decoration: underline;">Cell lines:</span></p>
<p>Human: A549, A673, BT-549, CD4 T, HCC1806, HeLa, HepG2, HFF, HK-GFP-MR, ILC, K562, KYSE-180, LapC4, M14, MCF7, MDA-MB-231, MDA-MB-436, RDES, SKNO1, VCaP, U2-OS, ZR-75-1 </p>
<p>Mouse: ESC, NPCs, BZ, GT1-7, acinar cells, HSPCs, Th2 cells, keratinocytes</p>
<p>Cattle: pbMEC, <span>MAC-T</span></p>
<p><span style="text-decoration: underline;">Tissues:</span></p>
<p>Mouse: kidney, heart, brain, iris, liver, limbs from E10.5 embryos</p>
<p><span>Horse: l</span>iver, brain, heart, lung, skeletal muscle, lamina, ovary</p>
<p><span style="text-decoration: underline;">ChIP on yeast</span></p>
<p>The iDeal ChIP-seq kit for TF is compatible with yeast samples. Check out our <strong><a href="https://www.diagenode.com/files/products/kits/Application_Note-ChIP_on_Yeast.pdf">Application Note</a></strong> presenting an optimized detailed protocol for ChIP on yeast.</p>
<p></p>
<p>Did you use the iDeal ChIP-seq for Transcription Factors Kit on other cell line / tissue / species? <a href="mailto:agnieszka.zelisko@diagenode.com?subject=Species, cell lines, tissues tested with the iDeal ChIP-seq Kit for TF&body=Dear Customer,%0D%0A%0D%0APlease, leave below your feedback about the iDeal ChIP-seq for Transcription Factors (cell / tissue type, species, other information...).%0D%0A%0D%0AThank you for sharing with us your experience !%0D%0A%0D%0ABest regards,%0D%0A%0D%0AAgnieszka Zelisko-Schmidt, PhD">Let us know!</a></p>',
'label3' => 'Additional solutions compatible with Auto iDeal ChIP-seq kit for Transcription Factors',
'info3' => '<p><span style="font-weight: 400;">The</span> <a href="https://www.diagenode.com/en/p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns"><span style="font-weight: 400;">Chromatin shearing optimization kit – Low SDS (iDeal Kit for TFs)</span></a><span style="font-weight: 400;"> is the kit compatible with the iDeal ChIP-seq kit for TF, recommended for the optimization of chromatin shearing, a critical step for ChIP.</span></p>
<p><a href="https://www.diagenode.com/en/p/chip-cross-link-gold-600-ul"><span style="font-weight: 400;">ChIP Cross-link Gold</span></a> <span style="font-weight: 400;">should be used in combination with formaldehyde when working with higher order and/or dynamic interactions, for efficient protein-protein fixation.</span></p>
<p><span style="font-weight: 400;">For library preparation of immunoprecipitated samples we recommend to use the </span><b> </b><a href="https://www.diagenode.com/en/categories/library-preparation-for-ChIP-seq"><span style="font-weight: 400;">MicroPlex Library Preparation Kit</span></a><span style="font-weight: 400;"> - validated for library preparation from picogram inputs.</span></p>
<p><a href="https://www.diagenode.com/en/categories/chip-seq-grade-antibodies"><span style="font-weight: 400;">ChIP-seq grade antibodies</span></a><span style="font-weight: 400;"> provide high yields with excellent specificity and sensitivity.</span></p>
<p><span style="font-weight: 400;">Check the list of available </span><a href="https://www.diagenode.com/en/categories/primer-pairs"><span style="font-weight: 400;">Primer pairs</span></a><span style="font-weight: 400;"> designed for high specificity to specific genomic regions.</span></p>',
'format' => '24 rxns',
'catalog_number' => 'C01010058',
'old_catalog_number' => '',
'sf_code' => 'C01010058-',
'type' => 'RFR',
'search_order' => '01-Accessory',
'price_EUR' => '915',
'price_USD' => '1130',
'price_GBP' => '840',
'price_JPY' => '143335',
'price_CNY' => '',
'price_AUD' => '2825',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns',
'meta_title' => 'Auto iDeal ChIP-seq Kit for Transcription Factors x24',
'meta_keywords' => '',
'meta_description' => 'Auto iDeal ChIP-seq Kit for Transcription Factors x24',
'modified' => '2021-11-23 10:51:46',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1923',
'antibody_id' => null,
'name' => 'MicroChIP DiaPure columns',
'description' => '<p><a href="https://www.diagenode.com/files/products/reagents/MicroChIP_DiaPure_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p>Diagenode’s MicroChIP DiaPure columns have been optimized for the purification and elution of very low amounts of DNA. This rapid method has been validated for epigenetic applications like low input ChIP (e.g. using the True MicroChIP kit) and CUT&Tag (e.g. using Diagenode’s pA-Tn5), but is also compatible with many other applications. The DNA can be eluted at high concentrations in volumes down to 6 μl and it is suitable for any downstream application (e.g. NGS).</p>
<p>Benefits of the MicroChIP DiaPure columns:</p>
<ul>
<li>Optimized for the purification of very low DNA amounts</li>
<li>Fast and easy protocol</li>
<li>Non-toxic</li>
<li>Validated for ChIP and Cut&Tag</li>
</ul>',
'label1' => 'Examples of results',
'info1' => '<h2 style="text-align: center;">MicroChIP DiaPure columns after ChIP</h2>
<p>Successful ChIP-seq results generated on 50,000 of K562 cells using True MicroChIP technology. ChIP has been performed accordingly to True MicroChIP protocol (Diagenode, Cat. No. C01010130), including DNA purification using the MicroChIP DiaPure columns. For the library preparation the MicroPlex Library Preparation Kit (Diagenode, Cat. No. C05010001) has been used. The below figure shows the peaks from ChIP-seq experiments using the following Diagenode antibodies: H3K4me1 (C15410194), H3K9/14ac (C15410200), H3K27ac (C15410196) and H3K36me3 (C15410192).</p>
<p><img src="https://www.diagenode.com/img/product/kits/figure-igv-microchip.png" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1:</strong> Integrative genomics viewer (IGV) visualization of ChIP-seq experiments using 50,000 of K562 cells.</p>
<p></p>
<h2 style="text-align: center;"></h2>
<h2 style="text-align: center;">MicroChIP DiaPure columns after CUT&Tag</h2>
<p>Successful CUT&Tag results showing a low background with high region-specific enrichment has been generated using 50.000 of K562 cells, 1 µg of H3K27me3 antibody (Diagenode, Cat. No. C15410069) and proteinA-Tn5 (1:250) (Diagenode, Cat. No. C01070001). 1 µg of IgG (Diagenode, Cat. No. C15410206) was used as negative control. Samples were purified using the MicroChIP DiaPure columns or phenol-chloroform purification. The below figure presenst the comparison of two purification methods.</p>
<p><img src="https://www.diagenode.com/img/product/kits/figure-diapure-igv.png" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 2:</strong> Integrative genomics viewer (IGV) visualization of CUT&Tag experiments: MicroChIP DiaPure columns vs phenol-chloroform purification using the H3K27me3 antibody.</p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C03040001',
'old_catalog_number' => '',
'sf_code' => 'C03040001-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'microchip-diapure-columns-50-rxns',
'meta_title' => 'MicroChIP DiaPure columns',
'meta_keywords' => '',
'meta_description' => 'MicroChIP DiaPure columns',
'modified' => '2023-04-20 16:11:26',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array([maximum depth reached])
),
(int) 4 => array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '9',
'position' => '10',
'parent_id' => '2',
'name' => 'ChIP-seq',
'description' => '<div class="row">
<div class="large-12 columns">Chromatin Immunoprecipitation (ChIP) coupled with high-throughput massively parallel sequencing as a detection method (ChIP-seq) has become one of the primary methods for epigenomics researchers, namely to investigate protein-DNA interaction on a genome-wide scale. This technique is now used in a variety of life science disciplines including cellular differentiation, tumor suppressor gene silencing, and the effect of histone modifications on gene expression.</div>
<div class="large-12 columns"></div>
<h5 class="large-12 columns"><strong></strong></h5>
<h5 class="large-12 columns"><strong>The ChIP-seq workflow</strong></h5>
<div class="small-12 medium-12 large-12 columns text-center"><br /><img src="https://www.diagenode.com/img/chip-seq-diagram.png" /></div>
<div class="large-12 columns"><br />
<ol>
<li class="large-12 columns"><strong>Chromatin preparation: </strong>Crosslink chromatin-bound proteins (histones or transcription factors) to DNA followed by cell lysis.</li>
<li class="large-12 columns"><strong>Chromatin shearing:</strong> Fragment chromatin by sonication to desired fragment size (100-500 bp)</li>
<li class="large-12 columns"><strong>Chromatin IP</strong>: Capture protein-DNA complexes with <strong><a href="../categories/chip-seq-grade-antibodies">specific ChIP-seq grade antibodies</a></strong> against the histone or transcription factor of interest</li>
<li class="large-12 columns"><strong>DNA purification</strong>: Reverse cross-links, elute, and purify </li>
<li class="large-12 columns"><strong>NGS Library Preparation</strong>: Ligate adapters and amplify IP'd material</li>
<li class="large-12 columns"><strong>Bioinformatic analysis</strong>: Perform r<span style="font-weight: 400;">ead filtering and trimming</span>, r<span style="font-weight: 400;">ead specific alignment, enrichment specific peak calling, QC metrics, multi-sample cross-comparison etc. </span></li>
</ol>
</div>
</div>
<div class="row" style="margin-top: 32px;">
<div class="small-12 medium-10 large-9 small-centered columns">
<div class="radius panel" style="background-color: #fff;">
<h3 class="text-center" style="color: #b21329;">Need guidance?</h3>
<p class="text-justify">Choose our full ChIP kits or simply choose what you need from antibodies, buffers, beads, chromatin shearing and purification reagents. With the ChIP Kit Customizer, you have complete flexibility on which components you want from our validated ChIP kits.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns"><a href="../pages/which-kit-to-choose"><img alt="" src="https://www.diagenode.com/img/banners/banner-decide.png" /></a></div>
<div class="small-6 medium-6 large-6 columns"><a href="../pages/chip-kit-customizer-1"><img alt="" src="https://www.diagenode.com/img/banners/banner-customizer.png" /></a></div>
</div>
</div>
</div>
</div>',
'in_footer' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'slug' => 'chromatin-immunoprecipitation-sequencing',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers wide range of kits and antibodies for Chromatin Immunoprecipitation Sequencing (ChIP-Seq) and also provides Bioruptor for chromatin shearing',
'meta_title' => 'Chromatin Immunoprecipitation - ChIP-seq Kits - Dna methylation | Diagenode',
'modified' => '2017-11-14 09:57:16',
'created' => '2015-04-12 18:08:46',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '117',
'position' => '3',
'parent_id' => '59',
'name' => 'Automated kits',
'description' => '',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'hide' => false,
'all_format' => false,
'is_antibody' => false,
'slug' => 'chromatin-ip-automated-kits',
'cookies_tag_id' => null,
'meta_keywords' => '',
'meta_description' => 'Diagenode’s validated ChIP-Seq Automated kits has everything you need for a successful ChIP prior to Next-Generation Sequencing.',
'meta_title' => 'Automated ChIP-Seq Kits for Chromatin Immunoprecipitation | Diagenode',
'modified' => '2022-02-11 14:11:01',
'created' => '2016-07-19 16:57:20',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '110',
'name' => 'Auto TrueMicroChIP kit',
'description' => '<div class="page" title="Page 4">
<div class="layoutArea">
<div class="column">
<p><span></span><span>Conventional ChIP protocols require high numbers of cells (hundreds of thousands cells at least) limiting the application for ChIP technology to few cell samples. Recently, ChIP assays on smallest amount of cells have been reported. The procedure requires tedious optimization of several reaction conditions to face the increased background observed in ChIP performed with reduced amount of cells. That might consequently lead to considerable time and lab expenditures. To reduce these tedious steps, Diagenode provides the new Auto True MicroChIP kit with optimized reagents and protocol to enable successful ChIP on as few as 10 000 cells. Moreover, the Auto True MicroChIP kit protocol has been thoroughly optimised by Diagenode for ChIP followed by high-throughput sequencing on Illumina</span><span>® </span><span>Next-Gen sequencers. To allow the generation of consistent results in ChIP-seq on 10 000 cells, Diagenode has also optimised a library preparation protocol on limited amount of immunoprecipitated DNA. </span></p>
<p><span>The new MicroPLEX library preparation kit allows the preparation of libraries for sequencing on picogram amount of immunoprecipitated DNA. Thus, association of the Auto True MicroChIP kit with the MicroPLEX library preparation kit provides optimised solutions to perform ChIP-sequencing on limited amount of cells. </span></p>
</div>
</div>
</div>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/Auto-TrueMicroChIP-kit-manual.pdf',
'slug' => 'auto-truemicrochip-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-09-01 12:30:58',
'created' => '2015-07-07 11:47:43',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '37',
'name' => 'Chromatin Brochure',
'description' => '<p>Whether you are experienced or new to the field of chromatin immunoprecipitation, Diagenode has everything you need to make ChIP easy and convenient while ensuring consistent data between samples and experiments. As an expert in the field of epigenetics, Diagenode is committed to providing complete solutions from chromatin shearing reagents, shearing instruments such as the Bioruptor® (the gold standard for chromatin shearing), ChIP kits, the largest number of validated and trusted antibodies on the market, and the SX-8G IP-Star® Compact Automated System to achieve unparalleled productivity and reproducibility.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf',
'slug' => 'chromatin-immunoprecipitation-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2022-03-24 12:34:11',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1775',
'name' => 'product/kits/chip-kit-icon.png',
'alt' => 'ChIP kit icon',
'modified' => '2018-04-17 11:52:29',
'created' => '2018-03-15 15:50:34',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '3563',
'name' => 'EZH2 is overexpressed in transitional preplasmablasts and is involved in human plasma cell differentiation.',
'authors' => 'Herviou L, Jourdan M, Martinez AM, Cavalli G, Moreaux J',
'description' => '<p>Plasma cells (PCs) play a major role in the defense of the host organism against pathogens. We have shown that PC generation can be modeled using multi-step culture systems that reproduce the sequential cell differentiation occurring in vivo. Using this unique model, we investigated the role of EZH2 during PC differentiation (PCD) using H3K27me3 and EZH2 ChIP-binding profiles. We then studied the effect of the inhibition of EZH2 enzymatic activity to understand how EZH2 regulates the key functions involved in PCD. EZH2 expression significantly increases in preplasmablasts with H3K27me3 mediated repression of genes involved in B cell and plasma cell identity. EZH2 was also found to be recruited to H3K27me3-free promoters of transcriptionally active genes known to regulate cell proliferation. Inhibition the catalytic activity of EZH2 resulted in B to PC transcriptional changes associated with PC maturation induction, as well as higher immunoglobulin secretion. Altogether, our data suggest that EZH2 is involved in the maintenance of preplasmablast transitory immature proliferative state that supports their amplification.</p>',
'date' => '2019-02-12',
'pmid' => 'http://www.pubmed.gov/30755708',
'doi' => '10.1038/s41375-019-0392-1',
'modified' => '2019-03-21 17:17:48',
'created' => '2019-03-21 14:12:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '3389',
'name' => 'An Integrated and Semiautomated Microscaled Approach to Profile Cis-Regulatory Elements by Histone Modification ChIP-Seq for Large-Scale Epigenetic Studies.',
'authors' => 'Youhanna Jankeel D, Cayford J, Schmiedel BJ, Vijayanand P, Seumois G.',
'description' => '<p>Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) is the preferred approach to map histone modifications and identify cis-regulatory DNA elements throughout the genome. Multiple methods have been described to increase the efficiency of library preparation and to reduce hands-on time as well as costs. This review describes detailed steps to perform cell fixation, chromatin shearing, immunoprecipitation, and sequencing library preparation for a batch of 48-96 samples with small cell numbers. The protocol implements a semiautomated platform to reduce technical variability and improve signal-to-noise ratio as well as reduce hands-on time, thus allowing large-scale epigenetic studies of clinical samples with limited cell numbers.</p>',
'date' => '2018-06-30',
'pmid' => 'http://www.pubmed.gov/29956160',
'doi' => '10.1007/978-1-4939-7896-0_22',
'modified' => '2018-11-09 12:29:33',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '3595',
'name' => 'Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche.',
'authors' => 'Baghdadi MB, Castel D, Machado L, Fukada SI, Birk DE, Relaix F, Tajbakhsh S, Mourikis P',
'description' => '<p>The cell microenvironment, which is critical for stem cell maintenance, contains both cellular and non-cellular components, including secreted growth factors and the extracellular matrix. Although Notch and other signalling pathways have previously been reported to regulate quiescence of stem cells, the composition and source of molecules that maintain the stem cell niche remain largely unknown. Here we show that adult muscle satellite (stem) cells in mice produce extracellular matrix collagens to maintain quiescence in a cell-autonomous manner. Using chromatin immunoprecipitation followed by sequencing, we identified NOTCH1/RBPJ-bound regulatory elements adjacent to specific collagen genes, the expression of which is deregulated in Notch-mutant mice. Moreover, we show that Collagen V (COLV) produced by satellite cells is a critical component of the quiescent niche, as depletion of COLV by conditional deletion of the Col5a1 gene leads to anomalous cell cycle entry and gradual diminution of the stem cell pool. Notably, the interaction of COLV with satellite cells is mediated by the Calcitonin receptor, for which COLV acts as a surrogate local ligand. Systemic administration of a calcitonin derivative is sufficient to rescue the quiescence and self-renewal defects found in COLV-null satellite cells. This study reveals a Notch-COLV-Calcitonin receptor signalling cascade that maintains satellite cells in a quiescent state in a cell-autonomous fashion, and raises the possibility that similar reciprocal mechanisms act in diverse stem cell populations.</p>',
'date' => '2018-05-23',
'pmid' => 'http://www.pubmed.gov/29795344',
'doi' => '10.1038/s41586-018-0144-9',
'modified' => '2019-04-17 15:12:55',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '3589',
'name' => 'A new metabolic gene signature in prostate cancer regulated by JMJD3 and EZH2.',
'authors' => 'Daures M, Idrissou M, Judes G, Rifaï K, Penault-Llorca F, Bignon YJ, Guy L, Bernard-Gallon D',
'description' => '<p>Histone methylation is essential for gene expression control. Trimethylated lysine 27 of histone 3 (H3K27me3) is controlled by the balance between the activities of JMJD3 demethylase and EZH2 methyltransferase. This epigenetic mark has been shown to be deregulated in prostate cancer, and evidence shows H3K27me3 enrichment on gene promoters in prostate cancer. To study the impact of this enrichment, a transcriptomic analysis with TaqMan Low Density Array (TLDA) of several genes was studied on prostate biopsies divided into three clinical grades: normal ( = 23) and two tumor groups that differed in their aggressiveness (Gleason score ≤ 7 ( = 20) and >7 ( = 19)). ANOVA demonstrated that expression of the gene set was upregulated in tumors and correlated with Gleason score, thus discriminating between the three clinical groups. Six genes involved in key cellular processes stood out: , , , , and . Chromatin immunoprecipitation demonstrated collocation of EZH2 and JMJD3 on gene promoters that was dependent on disease stage. Gene set expression was also evaluated on prostate cancer cell lines (DU 145, PC-3 and LNCaP) treated with an inhibitor of JMJD3 (GSK-J4) or EZH2 (DZNeP) to study their involvement in gene regulation. Results showed a difference in GSK-J4 sensitivity under PTEN status of cell lines and an opposite gene expression profile according to androgen status of cells. In summary, our data describe the impacts of JMJD3 and EZH2 on a new gene signature involved in prostate cancer that may help identify diagnostic and therapeutic targets in prostate cancer.</p>',
'date' => '2018-05-04',
'pmid' => 'http://www.pubmed.gov/29805743',
'doi' => '10.18632/oncotarget.25182',
'modified' => '2019-04-17 15:21:33',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '3193',
'name' => 'Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression',
'authors' => 'Ngollo M. et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">H3K27me3 histone marks shape the inhibition of gene transcription. In prostate cancer, the deregulation of H3K27me3 marks might play a role in prostate tumor progression.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">We investigated genome-wide H3K27me3 histone methylation profile using chromatin immunoprecipitation (ChIP) and 2X400K promoter microarrays to identify differentially-enriched regions in biopsy samples from prostate cancer patients. H3K27me3 marks were assessed in 34 prostate tumors: 11 with Gleason score > 7 (GS > 7), 10 with Gleason score ≤ 7 (GS ≤ 7), and 13 morphologically normal prostate samples.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">Here, H3K27me3 profiling identified an average of 386 enriched-genes on promoter regions in healthy control group versus 545 genes in GS ≤ 7 and 748 genes in GS > 7 group. We then ran a factorial discriminant analysis (FDA) and compared the enriched genes in prostate-tumor biopsies and normal biopsies using ANOVA to identify significantly differentially-enriched genes. The analysis identified ALG5, EXOSC8, CBX1, GRID2, GRIN3B, ING3, MYO1D, NPHP3-AS1, MSH6, FBXO11, SND1, SPATS2, TENM4 and TRA2A genes. These genes are possibly associated with prostate cancer. Notably, the H3K27me3 histone mark emerged as a novel regulatory mechanism in poor-prognosis prostate cancer.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Our findings point to epigenetic mark H3K27me3 as an important event in prostate carcinogenesis and progression. The results reported here provide new molecular insights into the pathogenesis of prostate cancer.</abstracttext></p>
</div>',
'date' => '2017-04-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28403887',
'doi' => '',
'modified' => '2017-06-19 14:07:35',
'created' => '2017-06-19 14:05:03',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array()
)
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array()
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010034</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1885" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1885" id="CartAdd/1885Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1885" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto hMeDIP kit x16 (monoclonal mouse antibody)</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns" data-reveal-id="cartModal-1885" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto hMeDIP kit x16 (monoclonal mouse antibody)</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-premium-bisulfite-kit-40-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02030031</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1893" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1893" id="CartAdd/1893Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1893" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto Premium Bisulfite kit</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto Premium Bisulfite kit',
'C02030031',
'240',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto Premium Bisulfite kit',
'C02030031',
'240',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-premium-bisulfite-kit-40-rxns" data-reveal-id="cartModal-1893" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto Premium Bisulfite kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns"><img src="/img/product/kits/chip-kit-icon.png" alt="ChIP kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C01010058</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1842" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1842" id="CartAdd/1842Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1842" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto iDeal ChIP-seq Kit for Transcription Factors</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto iDeal ChIP-seq Kit for Transcription Factors',
'C01010058',
'1130',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto iDeal ChIP-seq Kit for Transcription Factors',
'C01010058',
'1130',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns" data-reveal-id="cartModal-1842" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto iDeal ChIP-seq Kit for Transcription Factors</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/microchip-diapure-columns-50-rxns"><img src="/img/grey-logo.jpg" alt="default alt" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C03040001</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1923" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1923" id="CartAdd/1923Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1923" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MicroChIP DiaPure columns</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MicroChIP DiaPure columns',
'C03040001',
'125',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MicroChIP DiaPure columns',
'C03040001',
'125',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="microchip-diapure-columns-50-rxns" data-reveal-id="cartModal-1923" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MicroChIP DiaPure columns</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020011</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1888" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1888" id="CartAdd/1888Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1888" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto MethylCap kit</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1888" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto MethylCap kit</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '3624',
'product_id' => '1858',
'related_id' => '1888'
),
'Image' => array(
(int) 0 => array(
'id' => '1778',
'name' => 'product/kits/methyl-kit-icon.png',
'alt' => 'Methylation kit icon',
'modified' => '2019-04-23 15:17:01',
'created' => '2018-03-15 15:52:12',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$document = array(
'id' => '37',
'name' => 'Chromatin Brochure',
'description' => '<p>Whether you are experienced or new to the field of chromatin immunoprecipitation, Diagenode has everything you need to make ChIP easy and convenient while ensuring consistent data between samples and experiments. As an expert in the field of epigenetics, Diagenode is committed to providing complete solutions from chromatin shearing reagents, shearing instruments such as the Bioruptor® (the gold standard for chromatin shearing), ChIP kits, the largest number of validated and trusted antibodies on the market, and the SX-8G IP-Star® Compact Automated System to achieve unparalleled productivity and reproducibility.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf',
'slug' => 'chromatin-immunoprecipitation-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2022-03-24 12:34:11',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '1094',
'product_id' => '1858',
'document_id' => '37'
)
)
$publication = array(
'id' => '3193',
'name' => 'Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression',
'authors' => 'Ngollo M. et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">H3K27me3 histone marks shape the inhibition of gene transcription. In prostate cancer, the deregulation of H3K27me3 marks might play a role in prostate tumor progression.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">We investigated genome-wide H3K27me3 histone methylation profile using chromatin immunoprecipitation (ChIP) and 2X400K promoter microarrays to identify differentially-enriched regions in biopsy samples from prostate cancer patients. H3K27me3 marks were assessed in 34 prostate tumors: 11 with Gleason score > 7 (GS > 7), 10 with Gleason score ≤ 7 (GS ≤ 7), and 13 morphologically normal prostate samples.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">Here, H3K27me3 profiling identified an average of 386 enriched-genes on promoter regions in healthy control group versus 545 genes in GS ≤ 7 and 748 genes in GS > 7 group. We then ran a factorial discriminant analysis (FDA) and compared the enriched genes in prostate-tumor biopsies and normal biopsies using ANOVA to identify significantly differentially-enriched genes. The analysis identified ALG5, EXOSC8, CBX1, GRID2, GRIN3B, ING3, MYO1D, NPHP3-AS1, MSH6, FBXO11, SND1, SPATS2, TENM4 and TRA2A genes. These genes are possibly associated with prostate cancer. Notably, the H3K27me3 histone mark emerged as a novel regulatory mechanism in poor-prognosis prostate cancer.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Our findings point to epigenetic mark H3K27me3 as an important event in prostate carcinogenesis and progression. The results reported here provide new molecular insights into the pathogenesis of prostate cancer.</abstracttext></p>
</div>',
'date' => '2017-04-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28403887',
'doi' => '',
'modified' => '2017-06-19 14:07:35',
'created' => '2017-06-19 14:05:03',
'ProductsPublication' => array(
'id' => '2138',
'product_id' => '1858',
'publication_id' => '3193'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/28403887" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: campaign_id [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'carrousel_order' => array(
(int) 0 => 'ChIPmentation'
),
'language' => 'cn',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'meta_title' => 'Auto True MicroChIP kit',
'product' => array(
'Product' => array(
'id' => '1858',
'antibody_id' => null,
'name' => 'Auto True MicroChIP kit',
'description' => '<p>The Auto True MicroChIP kit in combination with the <a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns">MicroPlex Library Preparation™ kit</a> allows for performing ChIP-seq on as few as 10,000 cells. This microChIP-seq assay has been validated with the new generation <a href="../p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star<sup>®</sup> Compact Automated Workstation</a>. Furthermore Diagenode's high quality ChIP-seq grade antibodies have been used.</p>
<div class="row">
<div class="small-12 medium-5 large-6 columns"><small style="color: #666;">Video article</small>
<div class="flex-video"><iframe width="420" height="315" style="float: left; margin-right: 20px;" src="//www.youtube.com/embed/JZC9ARbLh_U?rel=0" frameborder="0" allowfullscreen="allowfullscreen"></iframe></div>
</div>
<div class="small-12 medium-7 large-6 columns">
<h3>Automating ChIP-seq Experiments to Generate Epigenetic Profiles on 10,000 HeLa Cells</h3>
<p><a href="https://www.youtube.com/watch?v=JZC9ARbLh_U"><img src="https://www.diagenode.com/img/jove_logo.png" alt="Jove logo" border="0" width="100" height="58" /></a></p>
</div>
</div>',
'label1' => '',
'info1' => '',
'label2' => 'Characteristics',
'info2' => '<ul>
<li><strong>Revolutionary:</strong> Only 10.000 cells needed for complete ChIP-seq procedure</li>
<li><strong>Reliable and accurate</strong> sequencing library preparation from just picogram inputs</li>
<li><strong>Validated on</strong> studies for histone marks and with the IP-Star<sup>®</sup> Compact Automated Platform</li>
</ul>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-histone-results.png" alt="ChIP on 10,000 cells" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>High efficiency ChIP on 10,000 cells</strong><br />ChIP efficiency on 10 000 cells. ChIP was performed on human Hela cells using the Diagenode antibodies H3K4me3 (Cat. No. pAb-003-050), H3K27ac (pAb-174-050), H3K9me3 (pAb-056-050) and H3K27me3 (pAb-069-050). Sheared chromatin from 10 000 cells and 0.1 µg (H3K27ac), 0.25 µg (H3K4me3 and H3K27me3) or 0.5 µg (H3K9me3) of the antibody were used per IP. Corresponding amount of IgG was used as control. Quantitative PCR was performed with primers for corresponding positive and negative loci. Figure shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-A.png" alt="ChIP amplification" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-B.png" alt="Match between True MicroChIP peaks and the reference dataset" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Reliable detection of enrichments in ChIP-seq</strong><br />A: ChIP has been peformed with H3K4me3 antibody, amplification of 17 pg of DNA ChIP'd from 10.000 cells and amplification of 35 pg of DNA ChIP'd from 100.000 cells (control experiment). The IP'd DNA was amplified and transformed into a sequencing-ready preparation for the Illumina plateform with the MicroPlex Library Preparation kit. The library was then analysed on an Illumina<sup>®</sup> Genome Analyzer. Cluster generation and sequencing were performed according to the manufacturer's instructions.<br />B: We observed a perfect match between the top 40% of True MicroChIP peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-result.png" alt="Cell lysis" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Hela cells were fixed with 1% formaldehyde (for 10 minutes at RT)</strong><br /> Cell lysis was performed using the Lysis Buffer tL1 of the Diagenode True MicroChIP kit. Samples corresponding to 10 000 cells are sheared during 5 rounds of 5 cycles of 30 seconds “ON” / 30 seconds “OFF” with the Bioruptor<sup>®</sup> Plus combined with the Bioruptor<sup>®</sup> Water cooler (Cat No. BioAcc-cool) at HIGH power setting (position H). For optimal results, samples are vortexed before and after performing 5 sonication cycles, followed by a short centrifugation at 4°C. 10 μl of DNA (equivalent to 60 000 cells) are analysed on a 1.5% agarose gel.</p>',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C01010140',
'old_catalog_number' => '',
'sf_code' => 'C01010140-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '540',
'price_USD' => '590',
'price_GBP' => '495',
'price_JPY' => '84590',
'price_CNY' => '',
'price_AUD' => '1475',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-true-microchip-kit-16-rxns',
'meta_title' => 'Auto True MicroChIP kit',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'modified' => '2019-05-10 09:28:26',
'created' => '2015-06-29 14:08:20',
'locale' => 'zho'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(),
'Group' => array(),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array()
)
)
$carrousel_order = array(
(int) 0 => 'ChIPmentation'
)
$language = 'cn'
$meta_keywords = ''
$meta_description = 'Auto True MicroChIP kit'
$meta_title = 'Auto True MicroChIP kit'
$product = array(
'Product' => array(
'id' => '1858',
'antibody_id' => null,
'name' => 'Auto True MicroChIP kit',
'description' => '<p>The Auto True MicroChIP kit in combination with the <a href="../p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns">MicroPlex Library Preparation™ kit</a> allows for performing ChIP-seq on as few as 10,000 cells. This microChIP-seq assay has been validated with the new generation <a href="../p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star<sup>®</sup> Compact Automated Workstation</a>. Furthermore Diagenode's high quality ChIP-seq grade antibodies have been used.</p>
<div class="row">
<div class="small-12 medium-5 large-6 columns"><small style="color: #666;">Video article</small>
<div class="flex-video"><iframe width="420" height="315" style="float: left; margin-right: 20px;" src="//www.youtube.com/embed/JZC9ARbLh_U?rel=0" frameborder="0" allowfullscreen="allowfullscreen"></iframe></div>
</div>
<div class="small-12 medium-7 large-6 columns">
<h3>Automating ChIP-seq Experiments to Generate Epigenetic Profiles on 10,000 HeLa Cells</h3>
<p><a href="https://www.youtube.com/watch?v=JZC9ARbLh_U"><img src="https://www.diagenode.com/img/jove_logo.png" alt="Jove logo" border="0" width="100" height="58" /></a></p>
</div>
</div>',
'label1' => '',
'info1' => '',
'label2' => 'Characteristics',
'info2' => '<ul>
<li><strong>Revolutionary:</strong> Only 10.000 cells needed for complete ChIP-seq procedure</li>
<li><strong>Reliable and accurate</strong> sequencing library preparation from just picogram inputs</li>
<li><strong>Validated on</strong> studies for histone marks and with the IP-Star<sup>®</sup> Compact Automated Platform</li>
</ul>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-histone-results.png" alt="ChIP on 10,000 cells" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>High efficiency ChIP on 10,000 cells</strong><br />ChIP efficiency on 10 000 cells. ChIP was performed on human Hela cells using the Diagenode antibodies H3K4me3 (Cat. No. pAb-003-050), H3K27ac (pAb-174-050), H3K9me3 (pAb-056-050) and H3K27me3 (pAb-069-050). Sheared chromatin from 10 000 cells and 0.1 µg (H3K27ac), 0.25 µg (H3K4me3 and H3K27me3) or 0.5 µg (H3K9me3) of the antibody were used per IP. Corresponding amount of IgG was used as control. Quantitative PCR was performed with primers for corresponding positive and negative loci. Figure shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-A.png" alt="ChIP amplification" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-reliable-B.png" alt="Match between True MicroChIP peaks and the reference dataset" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Reliable detection of enrichments in ChIP-seq</strong><br />A: ChIP has been peformed with H3K4me3 antibody, amplification of 17 pg of DNA ChIP'd from 10.000 cells and amplification of 35 pg of DNA ChIP'd from 100.000 cells (control experiment). The IP'd DNA was amplified and transformed into a sequencing-ready preparation for the Illumina plateform with the MicroPlex Library Preparation kit. The library was then analysed on an Illumina<sup>®</sup> Genome Analyzer. Cluster generation and sequencing were performed according to the manufacturer's instructions.<br />B: We observed a perfect match between the top 40% of True MicroChIP peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p><img src="https://www.diagenode.com/img/product/kits/true-micro-chip-result.png" alt="Cell lysis" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Hela cells were fixed with 1% formaldehyde (for 10 minutes at RT)</strong><br /> Cell lysis was performed using the Lysis Buffer tL1 of the Diagenode True MicroChIP kit. Samples corresponding to 10 000 cells are sheared during 5 rounds of 5 cycles of 30 seconds “ON” / 30 seconds “OFF” with the Bioruptor<sup>®</sup> Plus combined with the Bioruptor<sup>®</sup> Water cooler (Cat No. BioAcc-cool) at HIGH power setting (position H). For optimal results, samples are vortexed before and after performing 5 sonication cycles, followed by a short centrifugation at 4°C. 10 μl of DNA (equivalent to 60 000 cells) are analysed on a 1.5% agarose gel.</p>',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C01010140',
'old_catalog_number' => '',
'sf_code' => 'C01010140-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '540',
'price_USD' => '590',
'price_GBP' => '495',
'price_JPY' => '84590',
'price_CNY' => '',
'price_AUD' => '1475',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => true,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-true-microchip-kit-16-rxns',
'meta_title' => 'Auto True MicroChIP kit',
'meta_keywords' => '',
'meta_description' => 'Auto True MicroChIP kit',
'modified' => '2019-05-10 09:28:26',
'created' => '2015-06-29 14:08:20',
'locale' => 'zho'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(),
'Group' => array(),
'Related' => array(
(int) 0 => array(
'id' => '1885',
'antibody_id' => null,
'name' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'description' => '<p><span>The Auro hMeDIP kit is designed for enrichment of hydroxymethylated DNA from fragmented genomic DNA samples for use in genome-wide methylation analysis. It features</span><span> a highly specific monoclonal antibody against </span><span>5-hydroxymethylcytosine (5-hmC) for the immunoprecipitation of hydroxymethylated DNA</span><span>. It includes control DNA and primers to assess the effiency of the assay. </span><span>Performing hydroxymethylation profiling with the hMeDIP kit is fast, reliable and highly specific.</span></p>',
'label1' => ' Characteristics',
'info1' => '<ul style="list-style-type: disc;">
<li><span>Robust enrichment & immunoprecipitation of hydroxymethylated DNA</span></li>
<li>Highly specific monoclonal antibody against 5-hmC<span> for reliable, reproducible results</span></li>
<li>Including control DNA and primers to <span>monitor the efficiency of the assay</span>
<ul style="list-style-type: circle;">
<li>5-hmC, 5-mC and unmethylated DNA sequences and primer pairs</li>
<li>Mouse primer pairs against Sfi1 targeting hydroxymethylated gene in mouse</li>
</ul>
</li>
</ul>
<ul style="list-style-type: disc;">
<li>Improved single-tube, magnetic bead-based protocol</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '16 rxns',
'catalog_number' => 'C02010034',
'old_catalog_number' => '',
'sf_code' => 'C02010034-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '630',
'price_USD' => '690',
'price_GBP' => '580',
'price_JPY' => '98690',
'price_CNY' => '',
'price_AUD' => '1725',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns',
'meta_title' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'meta_keywords' => '',
'meta_description' => 'Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'modified' => '2021-01-18 10:37:19',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1893',
'antibody_id' => null,
'name' => 'Auto Premium Bisulfite kit',
'description' => '<p>Diagenode's Premium Bisulfite Kit rapidly converts DNA through bisulfite treatment. Our conversion reagent is added directly to DNA, requires no intermediate steps, and results in high yields of DNA ready for downstream analysis methods including PCR and Next-Generation Sequencing.</p>',
'label1' => 'Characteristics',
'info1' => '<p><strong>Bisulfite Conversion efficiency in TERTBS1 and CTS56 genomic regions.</strong></p>
<p>The figure shows the conversion efficiency in TERTBS1 and CTS56 genomic regions when using 1 μg, 500ng and 100 ng of genomic DNA. Between 1 and 5 clones were analyzed for the different amounts of genomic DNA</p>
<p><img src="https://www.diagenode.com/img/product/kits/auto-premium-bisulfite-clone.png" alt="Clone" /></p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '40 rxns',
'catalog_number' => 'C02030031',
'old_catalog_number' => '',
'sf_code' => 'C02030031-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '255',
'price_USD' => '240',
'price_GBP' => '230',
'price_JPY' => '39945',
'price_CNY' => '',
'price_AUD' => '600',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-premium-bisulfite-kit-40-rxns',
'meta_title' => 'Auto Premium Bisulfite kit',
'meta_keywords' => '',
'meta_description' => 'Auto Premium Bisulfite kit',
'modified' => '2019-12-16 10:05:26',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1842',
'antibody_id' => null,
'name' => 'Auto iDeal ChIP-seq Kit for Transcription Factors',
'description' => '<p><span><strong>This product must be used with the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star Compact Automated System</a>.</strong></span></p>
<p><span>Diagenode’s </span><strong>Auto iDeal ChIP-seq Kit for Transcription Factors</strong><span> is a highly specialized solution for robust Transcription Factor ChIP-seq results. Unlike competing solutions, our kit utilizes a highly optimized protocol and is backed by validation with a broad number and range of transcription factors. The kit provides high yields with excellent specificity and sensitivity.</span></p>',
'label1' => 'Characteristics',
'info1' => '<ul>
<li><strong>Confidence in results:</strong> Validated for ChIP-seq with multiple transcription factors</li>
<li><strong>Proven:</strong> Validated by the epigenetics community, including the BLUEPRINT consortium</li>
<li><strong>Most complete kit available</strong> for highest quality data - includes control antibodies and primers</li>
<li>Validated with Diagenode's <a href="https://www.diagenode.com/en/p/microplex-library-preparation-kit-v2-x12-12-indices-12-rxns"><span>MicroPlex Library Preparation™ kit</span></a> and <a href="https://www.diagenode.com/categories/ip-star" title="IP-Star Automated System">IP-Star<sup>®</sup></a> Automation System</li>
</ul>
<p> </p>
<h3>ChIP-seq on cells</h3>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-ctcf-diagenode.jpg" alt="CTCF Diagenode" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1.</strong> (A) Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina<sup>®</sup> HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the GAPDH positive control gene.</p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-figure-b-total-diagendoe-peaks.png" alt="CTCF Diagenode" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1B.</strong> The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>
<p> </p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-A.png" alt="ChIP-seq figure A" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-B.png" alt="ChIP-seq figure B" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-TF-chip-seq-C.png" alt="ChIP-seq figure C" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 2.</strong> Chromatin Immunoprecipitation has been performed using chromatin from HeLa cells, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade HDAC1 (A), LSD1 (B) and p53 antibody (C). The IP'd DNA was subsequently analysed on an Illumina<sup>®</sup> Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in regions of chromosome 3 (A), chromosome 12 (B) and chromosome 6 (C) respectively.</p>
<p> </p>
<h3>ChIP-seq on tissue</h3>
<p><img src="https://www.diagenode.com/img/product/kits/ideal-figure-3a.jpg" alt="ChIP-seq figure A" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 3A.</strong> Chromatin Immunoprecipitation has been performed using chromatin from mouse liver tissue, the iDeal ChIP-seq kit for Transcription Factors and the Diagenode ChIP-seq-grade CTCF antibody. The IP'd DNA was subsequently analysed on an Illumina® HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. This figure shows the peak distribution in a region surrounding the Vwf positive control gene.</p>
<p><img src="https://www.diagenode.com/img/product/kits/match-of-the-top40-peaks.png" alt="Match of the Top40 peaks" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 3B.</strong> The ChIP-seq dataset from this experiment has been compared with a reference dataset from the Broad Institute. We observed a perfect match between the top 40% of Diagenode peaks and the reference dataset. Based on the NIH Encode project criterion, ChIP-seq results are considered reproducible between an original and reproduced dataset if the top 40% of peaks have at least an 80% overlap ratio with the compared dataset.</p>',
'label2' => 'Species, cell lines, tissues tested',
'info2' => '<p>The iDeal ChIP-seq Kit for Transcription Factors is compatible with a broad variety of cell lines, tissues and species, as shown below. Other species / cell lines / tissues can be used with this kit.</p>
<p><span style="text-decoration: underline;">Cell lines:</span></p>
<p>Human: A549, A673, BT-549, CD4 T, HCC1806, HeLa, HepG2, HFF, HK-GFP-MR, ILC, K562, KYSE-180, LapC4, M14, MCF7, MDA-MB-231, MDA-MB-436, RDES, SKNO1, VCaP, U2-OS, ZR-75-1 </p>
<p>Mouse: ESC, NPCs, BZ, GT1-7, acinar cells, HSPCs, Th2 cells, keratinocytes</p>
<p>Cattle: pbMEC, <span>MAC-T</span></p>
<p><span style="text-decoration: underline;">Tissues:</span></p>
<p>Mouse: kidney, heart, brain, iris, liver, limbs from E10.5 embryos</p>
<p><span>Horse: l</span>iver, brain, heart, lung, skeletal muscle, lamina, ovary</p>
<p><span style="text-decoration: underline;">ChIP on yeast</span></p>
<p>The iDeal ChIP-seq kit for TF is compatible with yeast samples. Check out our <strong><a href="https://www.diagenode.com/files/products/kits/Application_Note-ChIP_on_Yeast.pdf">Application Note</a></strong> presenting an optimized detailed protocol for ChIP on yeast.</p>
<p></p>
<p>Did you use the iDeal ChIP-seq for Transcription Factors Kit on other cell line / tissue / species? <a href="mailto:agnieszka.zelisko@diagenode.com?subject=Species, cell lines, tissues tested with the iDeal ChIP-seq Kit for TF&body=Dear Customer,%0D%0A%0D%0APlease, leave below your feedback about the iDeal ChIP-seq for Transcription Factors (cell / tissue type, species, other information...).%0D%0A%0D%0AThank you for sharing with us your experience !%0D%0A%0D%0ABest regards,%0D%0A%0D%0AAgnieszka Zelisko-Schmidt, PhD">Let us know!</a></p>',
'label3' => 'Additional solutions compatible with Auto iDeal ChIP-seq kit for Transcription Factors',
'info3' => '<p><span style="font-weight: 400;">The</span> <a href="https://www.diagenode.com/en/p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns"><span style="font-weight: 400;">Chromatin shearing optimization kit – Low SDS (iDeal Kit for TFs)</span></a><span style="font-weight: 400;"> is the kit compatible with the iDeal ChIP-seq kit for TF, recommended for the optimization of chromatin shearing, a critical step for ChIP.</span></p>
<p><a href="https://www.diagenode.com/en/p/chip-cross-link-gold-600-ul"><span style="font-weight: 400;">ChIP Cross-link Gold</span></a> <span style="font-weight: 400;">should be used in combination with formaldehyde when working with higher order and/or dynamic interactions, for efficient protein-protein fixation.</span></p>
<p><span style="font-weight: 400;">For library preparation of immunoprecipitated samples we recommend to use the </span><b> </b><a href="https://www.diagenode.com/en/categories/library-preparation-for-ChIP-seq"><span style="font-weight: 400;">MicroPlex Library Preparation Kit</span></a><span style="font-weight: 400;"> - validated for library preparation from picogram inputs.</span></p>
<p><a href="https://www.diagenode.com/en/categories/chip-seq-grade-antibodies"><span style="font-weight: 400;">ChIP-seq grade antibodies</span></a><span style="font-weight: 400;"> provide high yields with excellent specificity and sensitivity.</span></p>
<p><span style="font-weight: 400;">Check the list of available </span><a href="https://www.diagenode.com/en/categories/primer-pairs"><span style="font-weight: 400;">Primer pairs</span></a><span style="font-weight: 400;"> designed for high specificity to specific genomic regions.</span></p>',
'format' => '24 rxns',
'catalog_number' => 'C01010058',
'old_catalog_number' => '',
'sf_code' => 'C01010058-',
'type' => 'RFR',
'search_order' => '01-Accessory',
'price_EUR' => '915',
'price_USD' => '1130',
'price_GBP' => '840',
'price_JPY' => '143335',
'price_CNY' => '',
'price_AUD' => '2825',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns',
'meta_title' => 'Auto iDeal ChIP-seq Kit for Transcription Factors x24',
'meta_keywords' => '',
'meta_description' => 'Auto iDeal ChIP-seq Kit for Transcription Factors x24',
'modified' => '2021-11-23 10:51:46',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1923',
'antibody_id' => null,
'name' => 'MicroChIP DiaPure columns',
'description' => '<p><a href="https://www.diagenode.com/files/products/reagents/MicroChIP_DiaPure_manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p>Diagenode’s MicroChIP DiaPure columns have been optimized for the purification and elution of very low amounts of DNA. This rapid method has been validated for epigenetic applications like low input ChIP (e.g. using the True MicroChIP kit) and CUT&Tag (e.g. using Diagenode’s pA-Tn5), but is also compatible with many other applications. The DNA can be eluted at high concentrations in volumes down to 6 μl and it is suitable for any downstream application (e.g. NGS).</p>
<p>Benefits of the MicroChIP DiaPure columns:</p>
<ul>
<li>Optimized for the purification of very low DNA amounts</li>
<li>Fast and easy protocol</li>
<li>Non-toxic</li>
<li>Validated for ChIP and Cut&Tag</li>
</ul>',
'label1' => 'Examples of results',
'info1' => '<h2 style="text-align: center;">MicroChIP DiaPure columns after ChIP</h2>
<p>Successful ChIP-seq results generated on 50,000 of K562 cells using True MicroChIP technology. ChIP has been performed accordingly to True MicroChIP protocol (Diagenode, Cat. No. C01010130), including DNA purification using the MicroChIP DiaPure columns. For the library preparation the MicroPlex Library Preparation Kit (Diagenode, Cat. No. C05010001) has been used. The below figure shows the peaks from ChIP-seq experiments using the following Diagenode antibodies: H3K4me1 (C15410194), H3K9/14ac (C15410200), H3K27ac (C15410196) and H3K36me3 (C15410192).</p>
<p><img src="https://www.diagenode.com/img/product/kits/figure-igv-microchip.png" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 1:</strong> Integrative genomics viewer (IGV) visualization of ChIP-seq experiments using 50,000 of K562 cells.</p>
<p></p>
<h2 style="text-align: center;"></h2>
<h2 style="text-align: center;">MicroChIP DiaPure columns after CUT&Tag</h2>
<p>Successful CUT&Tag results showing a low background with high region-specific enrichment has been generated using 50.000 of K562 cells, 1 µg of H3K27me3 antibody (Diagenode, Cat. No. C15410069) and proteinA-Tn5 (1:250) (Diagenode, Cat. No. C01070001). 1 µg of IgG (Diagenode, Cat. No. C15410206) was used as negative control. Samples were purified using the MicroChIP DiaPure columns or phenol-chloroform purification. The below figure presenst the comparison of two purification methods.</p>
<p><img src="https://www.diagenode.com/img/product/kits/figure-diapure-igv.png" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>Figure 2:</strong> Integrative genomics viewer (IGV) visualization of CUT&Tag experiments: MicroChIP DiaPure columns vs phenol-chloroform purification using the H3K27me3 antibody.</p>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '50 rxns',
'catalog_number' => 'C03040001',
'old_catalog_number' => '',
'sf_code' => 'C03040001-',
'type' => 'REF',
'search_order' => '04-undefined',
'price_EUR' => '125',
'price_USD' => '125',
'price_GBP' => '115',
'price_JPY' => '19580',
'price_CNY' => '',
'price_AUD' => '312',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'microchip-diapure-columns-50-rxns',
'meta_title' => 'MicroChIP DiaPure columns',
'meta_keywords' => '',
'meta_description' => 'MicroChIP DiaPure columns',
'modified' => '2023-04-20 16:11:26',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array([maximum depth reached])
),
(int) 4 => array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '9',
'position' => '10',
'parent_id' => '2',
'name' => 'ChIP-seq',
'description' => '<div class="row">
<div class="large-12 columns">Chromatin Immunoprecipitation (ChIP) coupled with high-throughput massively parallel sequencing as a detection method (ChIP-seq) has become one of the primary methods for epigenomics researchers, namely to investigate protein-DNA interaction on a genome-wide scale. This technique is now used in a variety of life science disciplines including cellular differentiation, tumor suppressor gene silencing, and the effect of histone modifications on gene expression.</div>
<div class="large-12 columns"></div>
<h5 class="large-12 columns"><strong></strong></h5>
<h5 class="large-12 columns"><strong>The ChIP-seq workflow</strong></h5>
<div class="small-12 medium-12 large-12 columns text-center"><br /><img src="https://www.diagenode.com/img/chip-seq-diagram.png" /></div>
<div class="large-12 columns"><br />
<ol>
<li class="large-12 columns"><strong>Chromatin preparation: </strong>Crosslink chromatin-bound proteins (histones or transcription factors) to DNA followed by cell lysis.</li>
<li class="large-12 columns"><strong>Chromatin shearing:</strong> Fragment chromatin by sonication to desired fragment size (100-500 bp)</li>
<li class="large-12 columns"><strong>Chromatin IP</strong>: Capture protein-DNA complexes with <strong><a href="../categories/chip-seq-grade-antibodies">specific ChIP-seq grade antibodies</a></strong> against the histone or transcription factor of interest</li>
<li class="large-12 columns"><strong>DNA purification</strong>: Reverse cross-links, elute, and purify </li>
<li class="large-12 columns"><strong>NGS Library Preparation</strong>: Ligate adapters and amplify IP'd material</li>
<li class="large-12 columns"><strong>Bioinformatic analysis</strong>: Perform r<span style="font-weight: 400;">ead filtering and trimming</span>, r<span style="font-weight: 400;">ead specific alignment, enrichment specific peak calling, QC metrics, multi-sample cross-comparison etc. </span></li>
</ol>
</div>
</div>
<div class="row" style="margin-top: 32px;">
<div class="small-12 medium-10 large-9 small-centered columns">
<div class="radius panel" style="background-color: #fff;">
<h3 class="text-center" style="color: #b21329;">Need guidance?</h3>
<p class="text-justify">Choose our full ChIP kits or simply choose what you need from antibodies, buffers, beads, chromatin shearing and purification reagents. With the ChIP Kit Customizer, you have complete flexibility on which components you want from our validated ChIP kits.</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns"><a href="../pages/which-kit-to-choose"><img alt="" src="https://www.diagenode.com/img/banners/banner-decide.png" /></a></div>
<div class="small-6 medium-6 large-6 columns"><a href="../pages/chip-kit-customizer-1"><img alt="" src="https://www.diagenode.com/img/banners/banner-customizer.png" /></a></div>
</div>
</div>
</div>
</div>',
'in_footer' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'slug' => 'chromatin-immunoprecipitation-sequencing',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers wide range of kits and antibodies for Chromatin Immunoprecipitation Sequencing (ChIP-Seq) and also provides Bioruptor for chromatin shearing',
'meta_title' => 'Chromatin Immunoprecipitation - ChIP-seq Kits - Dna methylation | Diagenode',
'modified' => '2017-11-14 09:57:16',
'created' => '2015-04-12 18:08:46',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '117',
'position' => '3',
'parent_id' => '59',
'name' => 'Automated kits',
'description' => '',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'hide' => false,
'all_format' => false,
'is_antibody' => false,
'slug' => 'chromatin-ip-automated-kits',
'cookies_tag_id' => null,
'meta_keywords' => '',
'meta_description' => 'Diagenode’s validated ChIP-Seq Automated kits has everything you need for a successful ChIP prior to Next-Generation Sequencing.',
'meta_title' => 'Automated ChIP-Seq Kits for Chromatin Immunoprecipitation | Diagenode',
'modified' => '2022-02-11 14:11:01',
'created' => '2016-07-19 16:57:20',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '110',
'name' => 'Auto TrueMicroChIP kit',
'description' => '<div class="page" title="Page 4">
<div class="layoutArea">
<div class="column">
<p><span></span><span>Conventional ChIP protocols require high numbers of cells (hundreds of thousands cells at least) limiting the application for ChIP technology to few cell samples. Recently, ChIP assays on smallest amount of cells have been reported. The procedure requires tedious optimization of several reaction conditions to face the increased background observed in ChIP performed with reduced amount of cells. That might consequently lead to considerable time and lab expenditures. To reduce these tedious steps, Diagenode provides the new Auto True MicroChIP kit with optimized reagents and protocol to enable successful ChIP on as few as 10 000 cells. Moreover, the Auto True MicroChIP kit protocol has been thoroughly optimised by Diagenode for ChIP followed by high-throughput sequencing on Illumina</span><span>® </span><span>Next-Gen sequencers. To allow the generation of consistent results in ChIP-seq on 10 000 cells, Diagenode has also optimised a library preparation protocol on limited amount of immunoprecipitated DNA. </span></p>
<p><span>The new MicroPLEX library preparation kit allows the preparation of libraries for sequencing on picogram amount of immunoprecipitated DNA. Thus, association of the Auto True MicroChIP kit with the MicroPLEX library preparation kit provides optimised solutions to perform ChIP-sequencing on limited amount of cells. </span></p>
</div>
</div>
</div>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/Auto-TrueMicroChIP-kit-manual.pdf',
'slug' => 'auto-truemicrochip-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-09-01 12:30:58',
'created' => '2015-07-07 11:47:43',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '37',
'name' => 'Chromatin Brochure',
'description' => '<p>Whether you are experienced or new to the field of chromatin immunoprecipitation, Diagenode has everything you need to make ChIP easy and convenient while ensuring consistent data between samples and experiments. As an expert in the field of epigenetics, Diagenode is committed to providing complete solutions from chromatin shearing reagents, shearing instruments such as the Bioruptor® (the gold standard for chromatin shearing), ChIP kits, the largest number of validated and trusted antibodies on the market, and the SX-8G IP-Star® Compact Automated System to achieve unparalleled productivity and reproducibility.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf',
'slug' => 'chromatin-immunoprecipitation-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2022-03-24 12:34:11',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1775',
'name' => 'product/kits/chip-kit-icon.png',
'alt' => 'ChIP kit icon',
'modified' => '2018-04-17 11:52:29',
'created' => '2018-03-15 15:50:34',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '3563',
'name' => 'EZH2 is overexpressed in transitional preplasmablasts and is involved in human plasma cell differentiation.',
'authors' => 'Herviou L, Jourdan M, Martinez AM, Cavalli G, Moreaux J',
'description' => '<p>Plasma cells (PCs) play a major role in the defense of the host organism against pathogens. We have shown that PC generation can be modeled using multi-step culture systems that reproduce the sequential cell differentiation occurring in vivo. Using this unique model, we investigated the role of EZH2 during PC differentiation (PCD) using H3K27me3 and EZH2 ChIP-binding profiles. We then studied the effect of the inhibition of EZH2 enzymatic activity to understand how EZH2 regulates the key functions involved in PCD. EZH2 expression significantly increases in preplasmablasts with H3K27me3 mediated repression of genes involved in B cell and plasma cell identity. EZH2 was also found to be recruited to H3K27me3-free promoters of transcriptionally active genes known to regulate cell proliferation. Inhibition the catalytic activity of EZH2 resulted in B to PC transcriptional changes associated with PC maturation induction, as well as higher immunoglobulin secretion. Altogether, our data suggest that EZH2 is involved in the maintenance of preplasmablast transitory immature proliferative state that supports their amplification.</p>',
'date' => '2019-02-12',
'pmid' => 'http://www.pubmed.gov/30755708',
'doi' => '10.1038/s41375-019-0392-1',
'modified' => '2019-03-21 17:17:48',
'created' => '2019-03-21 14:12:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '3389',
'name' => 'An Integrated and Semiautomated Microscaled Approach to Profile Cis-Regulatory Elements by Histone Modification ChIP-Seq for Large-Scale Epigenetic Studies.',
'authors' => 'Youhanna Jankeel D, Cayford J, Schmiedel BJ, Vijayanand P, Seumois G.',
'description' => '<p>Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) is the preferred approach to map histone modifications and identify cis-regulatory DNA elements throughout the genome. Multiple methods have been described to increase the efficiency of library preparation and to reduce hands-on time as well as costs. This review describes detailed steps to perform cell fixation, chromatin shearing, immunoprecipitation, and sequencing library preparation for a batch of 48-96 samples with small cell numbers. The protocol implements a semiautomated platform to reduce technical variability and improve signal-to-noise ratio as well as reduce hands-on time, thus allowing large-scale epigenetic studies of clinical samples with limited cell numbers.</p>',
'date' => '2018-06-30',
'pmid' => 'http://www.pubmed.gov/29956160',
'doi' => '10.1007/978-1-4939-7896-0_22',
'modified' => '2018-11-09 12:29:33',
'created' => '2018-11-08 12:59:45',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '3595',
'name' => 'Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche.',
'authors' => 'Baghdadi MB, Castel D, Machado L, Fukada SI, Birk DE, Relaix F, Tajbakhsh S, Mourikis P',
'description' => '<p>The cell microenvironment, which is critical for stem cell maintenance, contains both cellular and non-cellular components, including secreted growth factors and the extracellular matrix. Although Notch and other signalling pathways have previously been reported to regulate quiescence of stem cells, the composition and source of molecules that maintain the stem cell niche remain largely unknown. Here we show that adult muscle satellite (stem) cells in mice produce extracellular matrix collagens to maintain quiescence in a cell-autonomous manner. Using chromatin immunoprecipitation followed by sequencing, we identified NOTCH1/RBPJ-bound regulatory elements adjacent to specific collagen genes, the expression of which is deregulated in Notch-mutant mice. Moreover, we show that Collagen V (COLV) produced by satellite cells is a critical component of the quiescent niche, as depletion of COLV by conditional deletion of the Col5a1 gene leads to anomalous cell cycle entry and gradual diminution of the stem cell pool. Notably, the interaction of COLV with satellite cells is mediated by the Calcitonin receptor, for which COLV acts as a surrogate local ligand. Systemic administration of a calcitonin derivative is sufficient to rescue the quiescence and self-renewal defects found in COLV-null satellite cells. This study reveals a Notch-COLV-Calcitonin receptor signalling cascade that maintains satellite cells in a quiescent state in a cell-autonomous fashion, and raises the possibility that similar reciprocal mechanisms act in diverse stem cell populations.</p>',
'date' => '2018-05-23',
'pmid' => 'http://www.pubmed.gov/29795344',
'doi' => '10.1038/s41586-018-0144-9',
'modified' => '2019-04-17 15:12:55',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '3589',
'name' => 'A new metabolic gene signature in prostate cancer regulated by JMJD3 and EZH2.',
'authors' => 'Daures M, Idrissou M, Judes G, Rifaï K, Penault-Llorca F, Bignon YJ, Guy L, Bernard-Gallon D',
'description' => '<p>Histone methylation is essential for gene expression control. Trimethylated lysine 27 of histone 3 (H3K27me3) is controlled by the balance between the activities of JMJD3 demethylase and EZH2 methyltransferase. This epigenetic mark has been shown to be deregulated in prostate cancer, and evidence shows H3K27me3 enrichment on gene promoters in prostate cancer. To study the impact of this enrichment, a transcriptomic analysis with TaqMan Low Density Array (TLDA) of several genes was studied on prostate biopsies divided into three clinical grades: normal ( = 23) and two tumor groups that differed in their aggressiveness (Gleason score ≤ 7 ( = 20) and >7 ( = 19)). ANOVA demonstrated that expression of the gene set was upregulated in tumors and correlated with Gleason score, thus discriminating between the three clinical groups. Six genes involved in key cellular processes stood out: , , , , and . Chromatin immunoprecipitation demonstrated collocation of EZH2 and JMJD3 on gene promoters that was dependent on disease stage. Gene set expression was also evaluated on prostate cancer cell lines (DU 145, PC-3 and LNCaP) treated with an inhibitor of JMJD3 (GSK-J4) or EZH2 (DZNeP) to study their involvement in gene regulation. Results showed a difference in GSK-J4 sensitivity under PTEN status of cell lines and an opposite gene expression profile according to androgen status of cells. In summary, our data describe the impacts of JMJD3 and EZH2 on a new gene signature involved in prostate cancer that may help identify diagnostic and therapeutic targets in prostate cancer.</p>',
'date' => '2018-05-04',
'pmid' => 'http://www.pubmed.gov/29805743',
'doi' => '10.18632/oncotarget.25182',
'modified' => '2019-04-17 15:21:33',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '3193',
'name' => 'Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression',
'authors' => 'Ngollo M. et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">H3K27me3 histone marks shape the inhibition of gene transcription. In prostate cancer, the deregulation of H3K27me3 marks might play a role in prostate tumor progression.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">We investigated genome-wide H3K27me3 histone methylation profile using chromatin immunoprecipitation (ChIP) and 2X400K promoter microarrays to identify differentially-enriched regions in biopsy samples from prostate cancer patients. H3K27me3 marks were assessed in 34 prostate tumors: 11 with Gleason score > 7 (GS > 7), 10 with Gleason score ≤ 7 (GS ≤ 7), and 13 morphologically normal prostate samples.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">Here, H3K27me3 profiling identified an average of 386 enriched-genes on promoter regions in healthy control group versus 545 genes in GS ≤ 7 and 748 genes in GS > 7 group. We then ran a factorial discriminant analysis (FDA) and compared the enriched genes in prostate-tumor biopsies and normal biopsies using ANOVA to identify significantly differentially-enriched genes. The analysis identified ALG5, EXOSC8, CBX1, GRID2, GRIN3B, ING3, MYO1D, NPHP3-AS1, MSH6, FBXO11, SND1, SPATS2, TENM4 and TRA2A genes. These genes are possibly associated with prostate cancer. Notably, the H3K27me3 histone mark emerged as a novel regulatory mechanism in poor-prognosis prostate cancer.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Our findings point to epigenetic mark H3K27me3 as an important event in prostate carcinogenesis and progression. The results reported here provide new molecular insights into the pathogenesis of prostate cancer.</abstracttext></p>
</div>',
'date' => '2017-04-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28403887',
'doi' => '',
'modified' => '2017-06-19 14:07:35',
'created' => '2017-06-19 14:05:03',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array()
)
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array()
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02010034</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1885" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1885" id="CartAdd/1885Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1885" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto hMeDIP kit x16 (monoclonal mouse antibody)</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto hMeDIP kit x16 (monoclonal mouse antibody)',
'C02010034',
'690',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-hmedip-kit-x16-monoclonal-mouse-antibody-16-rxns" data-reveal-id="cartModal-1885" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto hMeDIP kit x16 (monoclonal mouse antibody)</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-premium-bisulfite-kit-40-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02030031</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1893" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1893" id="CartAdd/1893Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1893" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto Premium Bisulfite kit</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto Premium Bisulfite kit',
'C02030031',
'240',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto Premium Bisulfite kit',
'C02030031',
'240',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-premium-bisulfite-kit-40-rxns" data-reveal-id="cartModal-1893" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto Premium Bisulfite kit</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns"><img src="/img/product/kits/chip-kit-icon.png" alt="ChIP kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C01010058</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1842" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1842" id="CartAdd/1842Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1842" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto iDeal ChIP-seq Kit for Transcription Factors</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto iDeal ChIP-seq Kit for Transcription Factors',
'C01010058',
'1130',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto iDeal ChIP-seq Kit for Transcription Factors',
'C01010058',
'1130',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns" data-reveal-id="cartModal-1842" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto iDeal ChIP-seq Kit for Transcription Factors</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/microchip-diapure-columns-50-rxns"><img src="/img/grey-logo.jpg" alt="default alt" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C03040001</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1923" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1923" id="CartAdd/1923Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1923" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> MicroChIP DiaPure columns</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MicroChIP DiaPure columns',
'C03040001',
'125',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('MicroChIP DiaPure columns',
'C03040001',
'125',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="microchip-diapure-columns-50-rxns" data-reveal-id="cartModal-1923" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">MicroChIP DiaPure columns</h6>
</div>
</div>
</li>
<li>
<div class="row">
<div class="small-12 columns">
<a href="/cn/p/auto-methylcap-kit-x48-48-rxns"><img src="/img/product/kits/methyl-kit-icon.png" alt="Methylation kit icon" class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">C02020011</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
<!-- BEGIN: ADD TO CART MODAL --><div id="cartModal-1888" class="reveal-modal small" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<form action="/cn/carts/add/1888" id="CartAdd/1888Form" method="post" accept-charset="utf-8"><div style="display:none;"><input type="hidden" name="_method" value="POST"/></div><input type="hidden" name="data[Cart][product_id]" value="1888" id="CartProductId"/>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<p>将 <input name="data[Cart][quantity]" placeholder="1" value="1" min="1" style="width:60px;display:inline" type="number" id="CartQuantity" required="required"/> <strong> Auto MethylCap kit</strong> 添加至我的购物车。</p>
<div class="row">
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="checkout" id="checkout" value="checkout" type="submit">结账</button> </div>
<div class="small-6 medium-6 large-6 columns">
<button class="alert small button expand" onclick="$(this).addToCart('Auto MethylCap kit',
'C02020011',
'695',
$('#CartQuantity').val());" name="keepshop" id="keepshop" type="submit">继续购物</button> </div>
</div>
</div>
</div>
</form><a class="close-reveal-modal" aria-label="Close">×</a></div><!-- END: ADD TO CART MODAL --><a href="#" id="auto-methylcap-kit-x48-48-rxns" data-reveal-id="cartModal-1888" class="" style="color:#B21329"><i class="fa fa-cart-plus"></i></a>
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Auto MethylCap kit</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1888',
'antibody_id' => null,
'name' => 'Auto MethylCap kit',
'description' => '<p>The Auto MethylCap kit allows to specifically capture DNA fragments containing methylated CpGs. The assay is based on the affinity purification of methylated DNA using methyl-CpG-binding domain (MBD) of human MeCP2 protein. This procedure has been optimized to perform automated immunoprecipitation of chromatin using the <a href="https://www.diagenode.com/en/p/sx-8g-ip-star-compact-automated-system-1-unit">IP-Star® Compact Automated System</a> enabling highly reproducible results and allowing for high throughput.</p>',
'label1' => ' Characteristics',
'info1' => '<ul>
<li><strong>Fast & sensitive capture</strong> of methylated DNA</li>
<li><strong>High capture efficiency</strong></li>
<li><strong>Differential fractionation</strong> of methylated DNA by CpG density (3 eluted fractions)</li>
<li><strong>Automation compatibility</strong><strong></strong>
<h3>MBD-seq allows for detection of genomic regions with different CpG density</h3>
<p><img src="https://www.diagenode.com/img/product/kits/mbd_results1.png" alt="MBD-sequencing results have been validated by bisulfite sequencing" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p><strong>F</strong><strong>igure 1.</strong><span> </span>Using the MBD approach, two methylated regions were detected in different elution fractions according to their methylated CpG density (A). Low, Medium and High refer to the sequenced DNA from different elution fractions with increasing salt concentration. Methylated patterns of these two different methylated regions were validated by bisulfite conversion assay (B).<br /><strong></strong></p>
</li>
</ul>',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '48 rxns',
'catalog_number' => 'C02020011',
'old_catalog_number' => 'AF-Auto01-0048',
'sf_code' => 'C02020011-',
'type' => 'RFR',
'search_order' => '04-undefined',
'price_EUR' => '740',
'price_USD' => '695',
'price_GBP' => '675',
'price_JPY' => '115920',
'price_CNY' => '',
'price_AUD' => '1738',
'country' => 'ALL',
'except_countries' => 'Japan',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'auto-methylcap-kit-x48-48-rxns',
'meta_title' => 'Auto MethylCap kit x48',
'meta_keywords' => '',
'meta_description' => 'Auto MethylCap kit x48',
'modified' => '2020-09-17 13:33:12',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '3624',
'product_id' => '1858',
'related_id' => '1888'
),
'Image' => array(
(int) 0 => array(
'id' => '1778',
'name' => 'product/kits/methyl-kit-icon.png',
'alt' => 'Methylation kit icon',
'modified' => '2019-04-23 15:17:01',
'created' => '2018-03-15 15:52:12',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$document = array(
'id' => '37',
'name' => 'Chromatin Brochure',
'description' => '<p>Whether you are experienced or new to the field of chromatin immunoprecipitation, Diagenode has everything you need to make ChIP easy and convenient while ensuring consistent data between samples and experiments. As an expert in the field of epigenetics, Diagenode is committed to providing complete solutions from chromatin shearing reagents, shearing instruments such as the Bioruptor® (the gold standard for chromatin shearing), ChIP kits, the largest number of validated and trusted antibodies on the market, and the SX-8G IP-Star® Compact Automated System to achieve unparalleled productivity and reproducibility.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf',
'slug' => 'chromatin-immunoprecipitation-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2022-03-24 12:34:11',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '1094',
'product_id' => '1858',
'document_id' => '37'
)
)
$publication = array(
'id' => '3193',
'name' => 'Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression',
'authors' => 'Ngollo M. et al.',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">H3K27me3 histone marks shape the inhibition of gene transcription. In prostate cancer, the deregulation of H3K27me3 marks might play a role in prostate tumor progression.</abstracttext></p>
<h4>METHODS:</h4>
<p><abstracttext label="METHODS" nlmcategory="METHODS">We investigated genome-wide H3K27me3 histone methylation profile using chromatin immunoprecipitation (ChIP) and 2X400K promoter microarrays to identify differentially-enriched regions in biopsy samples from prostate cancer patients. H3K27me3 marks were assessed in 34 prostate tumors: 11 with Gleason score > 7 (GS > 7), 10 with Gleason score ≤ 7 (GS ≤ 7), and 13 morphologically normal prostate samples.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">Here, H3K27me3 profiling identified an average of 386 enriched-genes on promoter regions in healthy control group versus 545 genes in GS ≤ 7 and 748 genes in GS > 7 group. We then ran a factorial discriminant analysis (FDA) and compared the enriched genes in prostate-tumor biopsies and normal biopsies using ANOVA to identify significantly differentially-enriched genes. The analysis identified ALG5, EXOSC8, CBX1, GRID2, GRIN3B, ING3, MYO1D, NPHP3-AS1, MSH6, FBXO11, SND1, SPATS2, TENM4 and TRA2A genes. These genes are possibly associated with prostate cancer. Notably, the H3K27me3 histone mark emerged as a novel regulatory mechanism in poor-prognosis prostate cancer.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">Our findings point to epigenetic mark H3K27me3 as an important event in prostate carcinogenesis and progression. The results reported here provide new molecular insights into the pathogenesis of prostate cancer.</abstracttext></p>
</div>',
'date' => '2017-04-12',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/28403887',
'doi' => '',
'modified' => '2017-06-19 14:07:35',
'created' => '2017-06-19 14:05:03',
'ProductsPublication' => array(
'id' => '2138',
'product_id' => '1858',
'publication_id' => '3193'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/28403887" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
×