How to properly cite our product/service in your work We strongly recommend using this: CATS Small RNA-seq Kit x24 (Hologic Diagenode Cat# C05010040). Click here to copy to clipboard. Using our products or services in your publication? Let us know! |
Repeat RNAs associate with replication forks and post-replicative DNA.
Gylling HM, Gonzalez-Aguilera C, Smith MA, Kaczorowski DC, Groth A, Lund AH
Non-coding RNA has a proven ability to direct and regulate chromatin modifications by acting as scaffolds between DNA and histone-modifying complexes. However, it is unknown if ncRNA plays any role in DNA replication and epigenome maintenance, including histone eviction and re-instalment of histone-modifications aft... |
Multisite Evaluation of Next-Generation Methods for Small RNA Quantification.
Herbert ZT, Thimmapuram J, Xie S, Kershner JP, Kolling FW, Ringelberg CS, LeClerc A, Alekseyev YO, Fan J, Podnar JW, Stevenson HS, Sommerville G, Gupta S, Berkeley M, Koeman J, Perera A, Scott AR, Grenier JK, Malik J, Ashton JM, Pivarski KL, Wang X, Kuffe
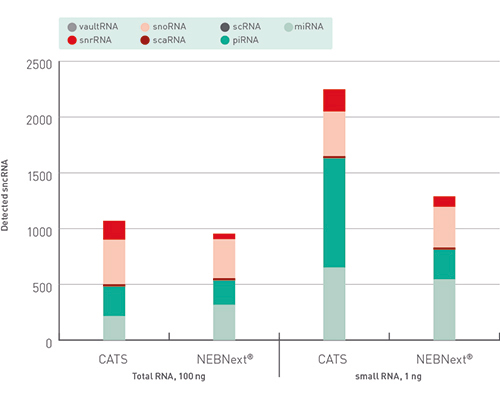
Small RNAs (smRNAs) are important regulators of many biologic processes and are now most frequently characterized using Illumina sequencing. However, although standard RNA sequencing library preparation has become routine in most sequencing facilities, smRNA sequencing library preparation has historically been chall... |
Circulating miRNA analysis for cancer diagnostics and therapy.
Valihrach L, Androvic P, Kubista M
Successful treatment of cancer depends on early diagnosis and effective monitoring of patients' response to therapy. Traditional tools based on tumor biopsies lack the sensitivity and specificity to capture cancer development in its early phases and are not applicable for continuous monitoring. To overcome these bar... |
The sncRNA Zoo: a repository for circulating small noncoding RNAs in animals.
Fehlmann T, Backes C, Pirritano M, Laufer T, Galata V, Kern F, Kahraman M, Gasparoni G, Ludwig N, Lenhof HP, Gregersen HA, Francke R, Meese E, Simon M, Keller A
The repertoire of small noncoding RNAs (sncRNAs), particularly miRNAs, in animals is considered to be evolutionarily conserved. Studies on sncRNAs are often largely based on homology-based information, relying on genomic sequence similarity and excluding actual expression data. To obtain information on sncRNA expres... |
MicroRNAs and Transplantation.
Khan Z, Suthanthiran M, Muthukumar T
miRNAs, ∼20 to 22 nucleotide single-stranded RNA species that play a pivotal role in the regulation of protein-coding genes, are emerging as robust biomarkers for assessing allograft status. Herein, the authors briefly review the biogenesis and function of the miRNAs and provide an overview of the tools to quant... |
NGS analysis of total small non coding RNAs from low input RNA from dried blood sampling
Marcello Pirritano, Tobias Fehlmann, Thomas Laufer, Nicole Ludwig, Gilles Gasparoni, Yongping Li, Eckart Meese, Andreas Keller, and Martin Simon
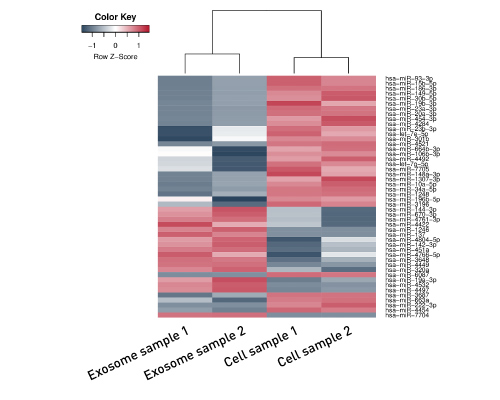
Circulating miRNAs are favored for biomarker candidates as they can reflect tissue specific miRNA dysregulation in disease contexts. Moreover, they have additional advantages that they can be monitored in a minimal invasive manner. Blood-borne miRNAs are therefore currently characterized to identify, describe and va... |