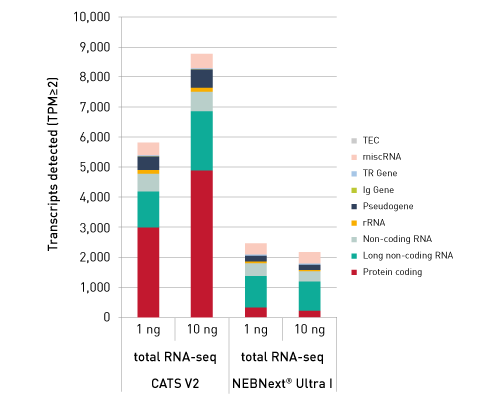
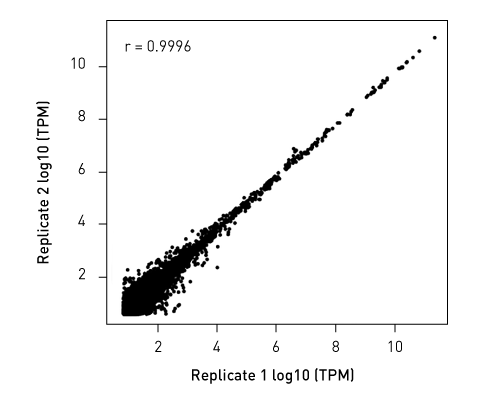
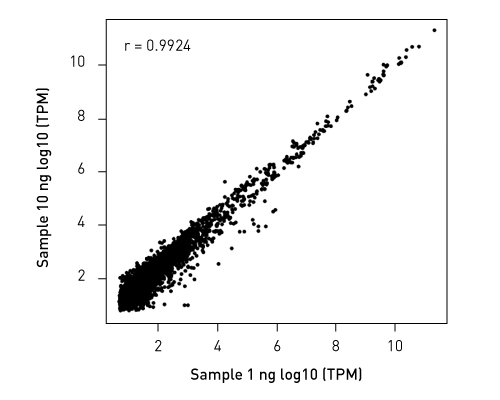
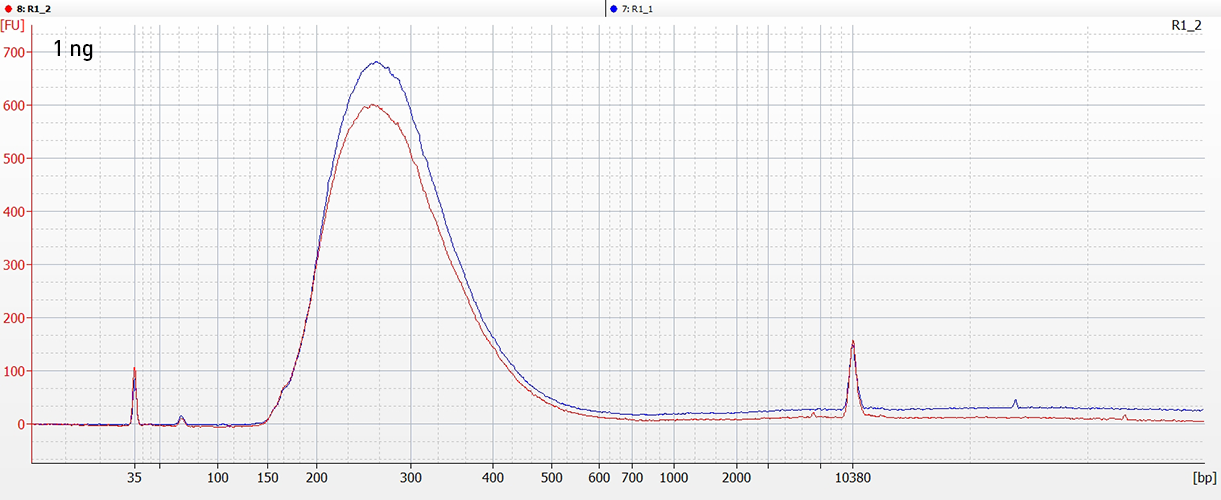
CATSは、RNAの3’末端をキャプチャーするためのポリヌクレオチドテーリングおよび、cDNA合成時に5’末端をキャプチャーするためのMMLV-RTによるテンプレートスイッチ 能力を利用した技術です。この方法なら、rRNAが 除去済のサンプルを使用の場合、100ピコグラムの低インプットRNA量からわずか数時間でIllumina用シークエンシングライブラリーを作製することができます。
ライブラリーは、ストランド特異的な情報を保持しています。「CATS RNA-seq Kit」は、増幅を最小限に抑えることでより多くの有効な鋳型をキャプチャーし、また、バイアスを最小限に抑えることで高い複雑性を得ることができるので、他のライゲーションベースの手法と比べより高いライブラリー作製効率を示します。