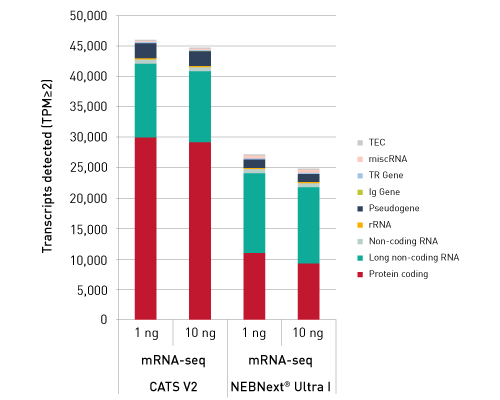
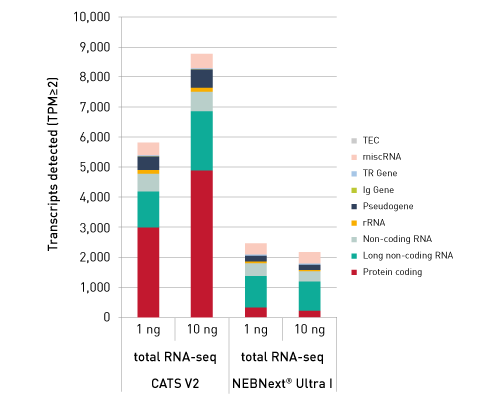
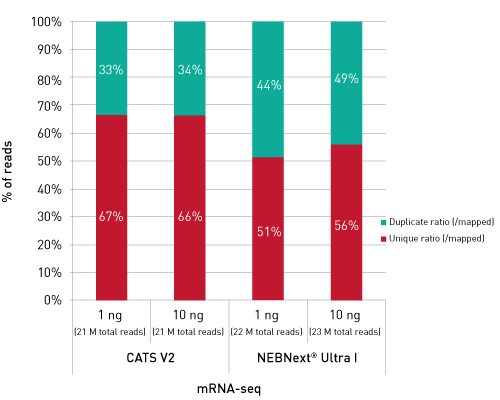
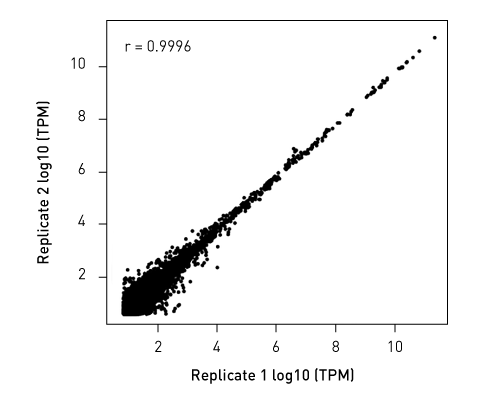
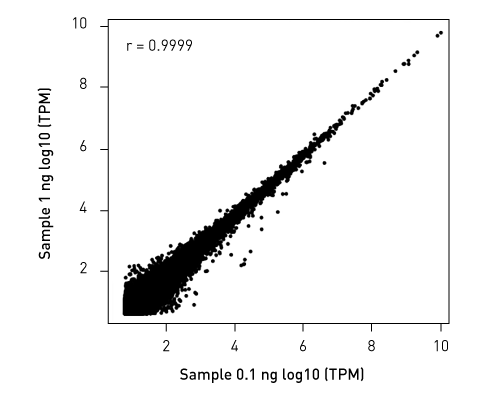
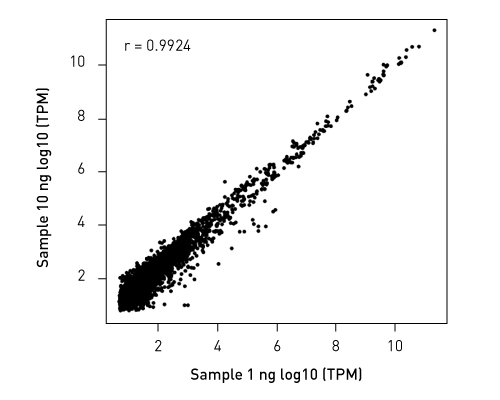
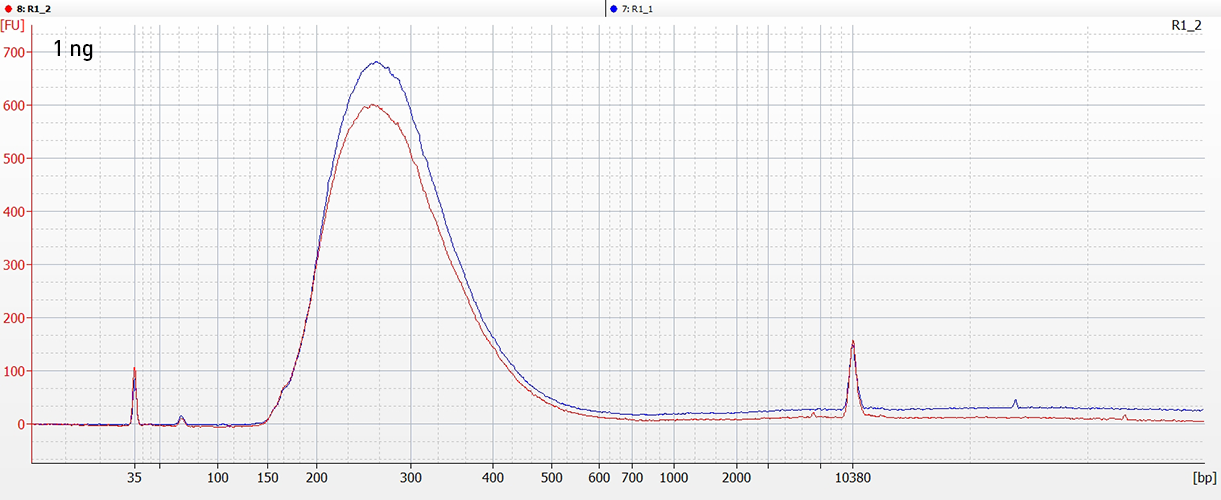
CATSで核酸3末端を補足する際のポリヌクレオチドテーリング、またはcDNA合成する5'末端を捕捉する際のMMLV-RTのテンプレートスイッチング能力が非常に重要になります。このキットなら、わずか数時間で10ピコグラム以下のRNA入力からIllumina用互換のシーケンシング準備済ライブラリーを作成することができます。.
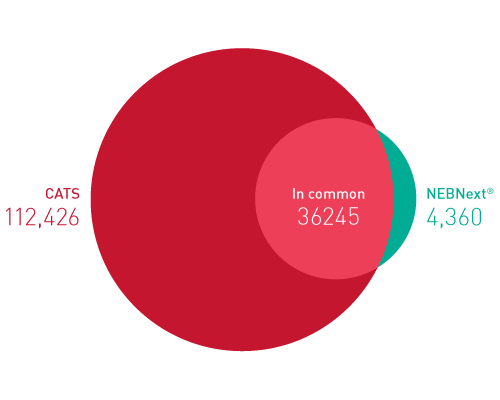
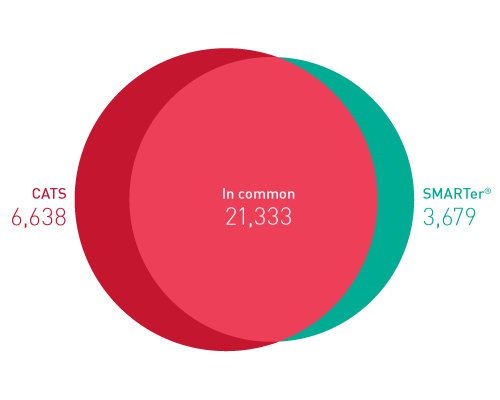
ライブラリーは、起源の鎖特異性を保持します。他社のソリューションとは異なり、当社のCATS RNA-seqキットはライゲーションベースの方法と比較してもより高いライブラリー効率を示し、より良いテンプレート捕捉からさらに高い複雑性、より少ない増幅条件による最小バイアスをご提供します。