Notice (8): Undefined variable: solution_of_interest [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'product' => array(
'Product' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
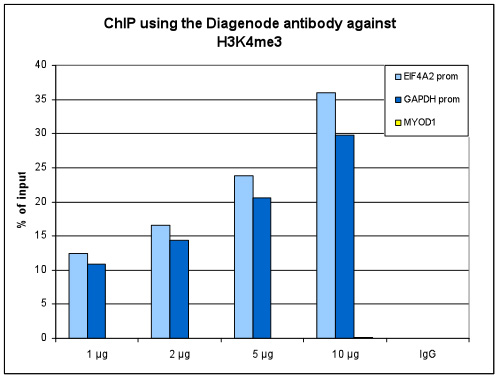
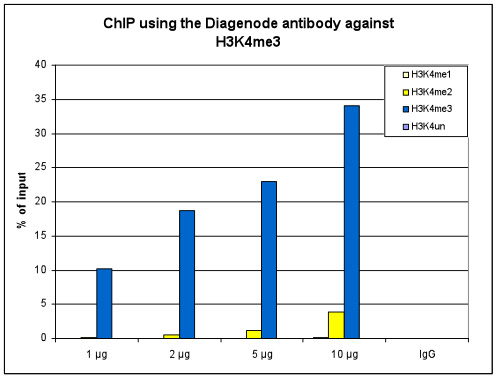
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
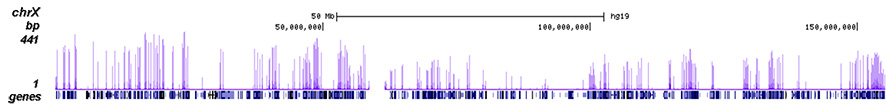
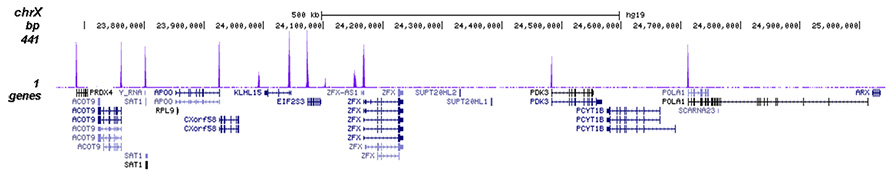
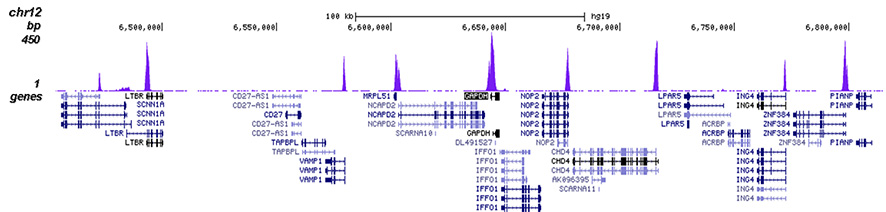
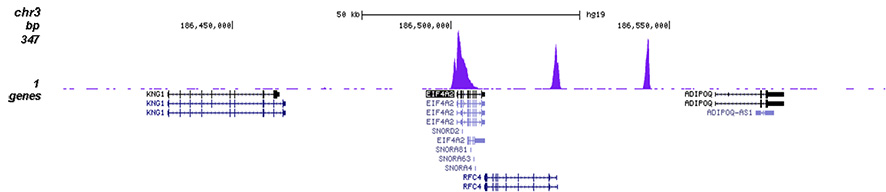
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
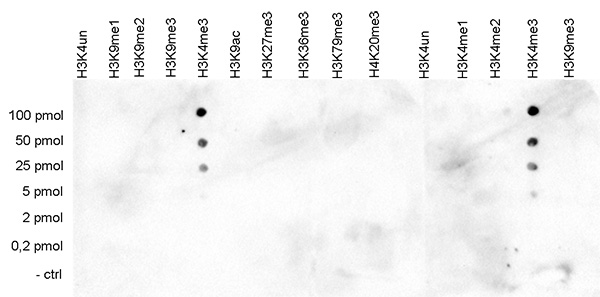
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
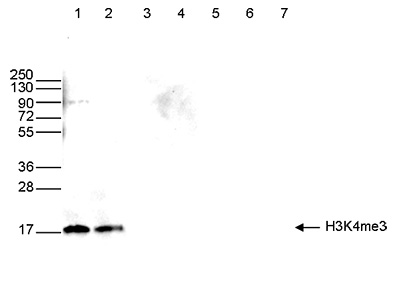
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
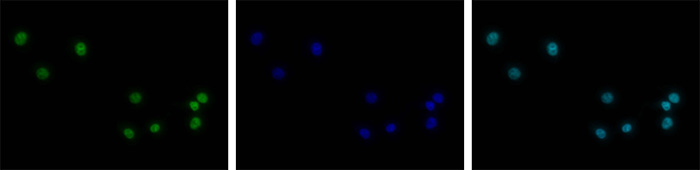
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '114',
'name' => 'H3K4me3 polyclonal antibody',
'description' => 'Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.',
'clonality' => '',
'isotype' => '',
'lot' => '002',
'concentration' => '1.4 µg/µl',
'reactivity' => 'Human, mouse, Arabidopsis: positive. Other species: not tested.',
'type' => 'Polyclonal',
'purity' => 'Affinity purified polyclonal antibody.',
'classification' => 'Classic',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>ChIP/ChIP-seq <sup>*</sup></td>
<td>1 µg/ChIP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting</td>
<td>1:2,000</td>
<td>Fig 3</td>
</tr>
<tr>
<td>Western Blotting</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
<tr>
<td>Immunofluorescence</td>
<td>1:100</td>
<td>Fig 5</td>
</tr>
</tbody>
</table>
<p></p>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 µg per IP.</small></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide and 0.05% ProClin 300.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-11-25 17:04:17',
'created' => '0000-00-00 00:00:00',
'select_label' => '114 - H3K4me3 polyclonal antibody (002 - 1.4 µg/µl - Human, mouse, Arabidopsis: positive. Other species: not tested. - Affinity purified polyclonal antibody. - Rabbit)'
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/h3k4me3-polyclonal-antibody-classic-50-ug-25-ul'
)
$language = 'en'
$meta_keywords = 'H3K4me3 Antibody '
$meta_description = 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.'
$meta_title = 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode'
$product = array(
'Product' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '114',
'name' => 'H3K4me3 polyclonal antibody',
'description' => 'Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.',
'clonality' => '',
'isotype' => '',
'lot' => '002',
'concentration' => '1.4 µg/µl',
'reactivity' => 'Human, mouse, Arabidopsis: positive. Other species: not tested.',
'type' => 'Polyclonal',
'purity' => 'Affinity purified polyclonal antibody.',
'classification' => 'Classic',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>ChIP/ChIP-seq <sup>*</sup></td>
<td>1 µg/ChIP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting</td>
<td>1:2,000</td>
<td>Fig 3</td>
</tr>
<tr>
<td>Western Blotting</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
<tr>
<td>Immunofluorescence</td>
<td>1:100</td>
<td>Fig 5</td>
</tr>
</tbody>
</table>
<p></p>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 µg per IP.</small></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide and 0.05% ProClin 300.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-11-25 17:04:17',
'created' => '0000-00-00 00:00:00',
'select_label' => '114 - H3K4me3 polyclonal antibody (002 - 1.4 µg/µl - Human, mouse, Arabidopsis: positive. Other species: not tested. - Affinity purified polyclonal antibody. - Rabbit)'
),
'Slave' => array(
(int) 0 => array(
'id' => '322',
'name' => 'C15410030',
'product_id' => '2195',
'modified' => '2020-11-25 16:06:48',
'created' => '2020-11-25 16:06:48'
)
),
'Group' => array(
'Group' => array(
'id' => '322',
'name' => 'C15410030',
'product_id' => '2195',
'modified' => '2020-11-25 16:06:48',
'created' => '2020-11-25 16:06:48'
),
'Master' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '28',
'position' => '10',
'parent_id' => '40',
'name' => 'DB',
'description' => '<p>Dot blotting</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'dot-blotting',
'meta_keywords' => 'Dot blotting,Monoclonal & Polyclonal antibody,',
'meta_description' => 'Diagenode offers Monoclonal & Polyclonal antibodies for Dot blotting applications',
'meta_title' => 'Dot blotting Antibodies - Monoclonal & Polyclonal antibody | Diagenode',
'modified' => '2016-01-13 14:40:49',
'created' => '2015-07-08 13:45:05',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '19',
'position' => '10',
'parent_id' => '40',
'name' => 'WB',
'description' => '<p><strong>Western blot</strong> : The quality of antibodies used in this technique is crucial for correct and specific protein identification. Diagenode offers huge selection of highly sensitive and specific western blot-validated antibodies.</p>
<p>Learn more about: <a href="https://www.diagenode.com/applications/western-blot">Loading control, MW marker visualization</a><em>. <br /></em></p>
<p><em></em>Check our selection of antibodies validated in Western blot.</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'western-blot-antibodies',
'meta_keywords' => ' Western Blot Antibodies ,western blot protocol,Western Blotting Products,Polyclonal antibodies ,monoclonal antibodies ',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for western blot applications',
'meta_title' => ' Western Blot - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-26 12:44:51',
'created' => '2015-01-07 09:20:00',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '111',
'position' => '40',
'parent_id' => '4',
'name' => 'Histone antibodies',
'description' => '<p>Histones are the main protein components of chromatin involved in the compaction of DNA into nucleosomes, the basic units of chromatin. A <strong>nucleosome</strong> consists of one pair of each of the core histones (<strong>H2A</strong>, <strong>H2B</strong>, <strong>H3</strong> and <strong>H4</strong>) forming an octameric structure wrapped by 146 base pairs of DNA. The different nucleosomes are linked by the linker histone<strong> H1, </strong>allowing for further condensation of chromatin.</p>
<p>The core histones have a globular structure with large unstructured N-terminal tails protruding from the nucleosome. They can undergo to multiple post-translational modifications (PTM), mainly at the N-terminal tails. These <strong>post-translational modifications </strong>include methylation, acetylation, phosphorylation, ubiquitinylation, citrullination, sumoylation, deamination and crotonylation. The most well characterized PTMs are <strong>methylation,</strong> <strong>acetylation and phosphorylation</strong>. Histone methylation occurs mainly on lysine (K) residues, which can be mono-, di- or tri-methylated, and on arginines (R), which can be mono-methylated and symmetrically or asymmetrically di-methylated. Histone acetylation occurs on lysines and histone phosphorylation mainly on serines (S), threonines (T) and tyrosines (Y).</p>
<p>The PTMs of the different residues are involved in numerous processes such as DNA repair, DNA replication and chromosome condensation. They influence the chromatin organization and can be positively or negatively associated with gene expression. Trimethylation of H3K4, H3K36 and H3K79, and lysine acetylation generally result in an open chromatin configuration (figure below) and are therefore associated with <strong>euchromatin</strong> and gene activation. Trimethylation of H3K9, K3K27 and H4K20, on the other hand, is enriched in <strong>heterochromatin </strong>and associated with gene silencing. The combination of different histone modifications is called the "<strong>histone code</strong>”, analogous to the genetic code.</p>
<p><img src="https://www.diagenode.com/img/categories/antibodies/histone-marks-illustration.png" /></p>
<p>Diagenode is proud to offer a large range of antibodies against histones and histone modifications. Our antibodies are highly specific and have been validated in many applications, including <strong>ChIP</strong> and <strong>ChIP-seq</strong>.</p>
<p>Diagenode’s collection includes antibodies recognizing:</p>
<ul>
<li><strong>Histone H1 variants</strong></li>
<li><strong>Histone H2A, H2A variants and histone H2A</strong> <strong>modifications</strong> (serine phosphorylation, lysine acetylation, lysine ubiquitinylation)</li>
<li><strong>Histone H2B and H2B</strong> <strong>modifications </strong>(serine phosphorylation, lysine acetylation)</li>
<li><strong>Histone H3 and H3 modifications </strong>(lysine methylation (mono-, di- and tri-methylated), lysine acetylation, serine phosphorylation, threonine phosphorylation, arginine methylation (mono-methylated, symmetrically and asymmetrically di-methylated))</li>
<li><strong>Histone H4 and H4 modifications (</strong>lysine methylation (mono-, di- and tri-methylated), lysine acetylation, arginine methylation (mono-methylated and symmetrically di-methylated), serine phosphorylation )</li>
</ul>
<p><span style="font-weight: 400;"><strong>HDAC's HAT's, HMT's and other</strong> <strong>enzymes</strong> which modify histones can be found in the category <a href="../categories/chromatin-modifying-proteins-histone-transferase">Histone modifying enzymes</a><br /></span></p>
<p><span style="font-weight: 400;"> Diagenode’s highly validated antibodies:</span></p>
<ul>
<li><span style="font-weight: 400;"> Highly sensitive and specific</span></li>
<li><span style="font-weight: 400;"> Cost-effective (requires less antibody per reaction)</span></li>
<li><span style="font-weight: 400;"> Batch-specific data is available on the website</span></li>
<li><span style="font-weight: 400;"> Expert technical support</span></li>
<li><span style="font-weight: 400;"> Sample sizes available</span></li>
<li><span style="font-weight: 400;"> 100% satisfaction guarantee</span></li>
</ul>',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'histone-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Histone, antibody, histone h1, histone h2, histone h3, histone h4',
'meta_description' => 'Polyclonal and Monoclonal Antibodies against Histones and their modifications validated for many applications, including Chromatin Immunoprecipitation (ChIP) and ChIP-Sequencing (ChIP-seq)',
'meta_title' => 'Histone and Modified Histone Antibodies | Diagenode',
'modified' => '2020-09-17 13:34:56',
'created' => '2016-04-01 16:01:32',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '127',
'position' => '10',
'parent_id' => '4',
'name' => 'ChIP-grade antibodies',
'description' => '<div class="row">
<div class="small-10 columns"><center></center>
<p><br />Chromatin immunoprecipitation (<b>ChIP</b>) is a technique to study the associations of proteins with the specific genomic regions in intact cells. One of the most important steps of this protocol is the immunoprecipitation of targeted protein using the antibody specifically recognizing it. The quality of antibodies used in ChIP is essential for the success of the experiment. Diagenode offers extensively validated ChIP-grade antibodies, confirmed for their specificity, and high level of performance in ChIP. Each batch is validated, and batch-specific data are available on the website.</p>
<p></p>
</div>
<div class="small-2 columns"><img src="https://www.diagenode.com/emailing/images/epi-success-guaranteed-icon.png" alt="Epigenetic success guaranteed" /></div>
</div>
<p><strong>ChIP results</strong> obtained with the antibody directed against H3K4me3 (Cat. No. <a href="../p/h3k4me3-polyclonal-antibody-premium-50-ug-50-ul">C15410003</a>). </p>
<div class="row">
<div class="small-12 medium-6 large-6 columns"><img src="https://www.diagenode.com/img/product/antibodies/C15410003-fig1-ChIP.jpg" alt="" width="400" height="315" /> </div>
<div class="small-12 medium-6 large-6 columns">
<p></p>
<p></p>
<p></p>
</div>
</div>
<p></p>
<p>Our aim at Diagenode is to offer the largest collection of highly specific <strong>ChIP-grade antibodies</strong>. We add new antibodies monthly. Find your ChIP-grade antibody in the list below and check more information about tested applications, extensive validation data, and product information.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'chip-grade-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'ChIP-grade antibodies, polyclonal antibody, monoclonal antibody, Diagenode',
'meta_description' => 'Diagenode Offers Extensively Validated ChIP-Grade Antibodies, Confirmed for their Specificity, and high level of Performance in Chromatin Immunoprecipitation ChIP',
'meta_title' => 'Chromatin immunoprecipitation ChIP-grade antibodies | Diagenode',
'modified' => '2021-07-01 10:22:38',
'created' => '2017-02-14 11:16:04',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '405',
'name' => 'Datasheet H3K4me3 C15410030',
'description' => '<p>Datasheet description</p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_H3K4me3_C15410030.pdf',
'slug' => 'datasheet-h3k4me3-C15410030',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-11-20 17:40:11',
'created' => '2015-07-07 11:47:44',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1783',
'name' => 'product/antibodies/chipseq-grade-ab-icon.png',
'alt' => 'ChIP-seq Grade',
'modified' => '2020-11-27 07:04:40',
'created' => '2018-03-15 15:54:09',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4598',
'name' => 'SUMO protease FUG1, histone reader AL3 and the PRC1 Complex areintegral to repeat-expansion induced epigenetic silencing in Arabidopsisthaliana',
'authors' => 'Sureshkumar S. et al.',
'description' => '<p>Epigenetic gene silencing induced by expanded repeats can cause diverse phenotypes ranging from severe growth defects in plants to genetic diseases such as Friedreich’s ataxia in humans1. The molecular mechanisms underlying repeat expansion-induced epigenetic silencing remain largely unknown2,3. Using a plant model, we have previously shown that expanded repeats can induce smallRNAs which in turn can lead to epigenetic silencing through the RNA-dependent DNA methylation pathway4,5. Here, using a genetic suppressor screen, we confirm a key role for the RdDM pathway and identify novel components required for epigenetic silencing caused by expanded repeats. We show that FOURTH ULP LIKE GENE CLASS 1 (FUG1) – a SUMO protease, ALFIN-LIKE 3 – a histone reader and LIKE HETEROCHROMATIN 1 (LHP1) - a component of the PRC1 complex are required for repeat expansion-induced epigenetic silencing. Loss of any of these components suppress repeat expansion-associated phenotypes. SUMO protease FUG1 physically interacts with AL3 and perturbing its potential SUMOylation site disrupts its nuclear localisation. AL3 physically interacts with LHP1 of the PRC1 complex and the FUG1-AL3-LHP1 module is essential to confer repeat expansion-associated epigenetic silencing. Our findings highlight the importance post-translational modifiers and histone readers in epigenetic silencing caused by repeat expansions.</p>',
'date' => '2023-12-01',
'pmid' => 'https://doi.org/10.1101%2F2023.01.13.523841',
'doi' => '10.1101/2023.01.13.523841',
'modified' => '2023-04-06 09:10:33',
'created' => '2023-02-21 09:59:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4692',
'name' => 'Temporal modification of H3K9/14ac and H3K4me3 histone marksmediates mechano-responsive gene expression during the accommodationprocess in poplar',
'authors' => 'Ghosh R. et al.',
'description' => '<p>Plants can attenuate their molecular response to repetitive mechanical stimulation as a function of their mechanical history. For instance, a single bending of stem is sufficient to attenuate the gene expression in poplar plants to the subsequent mechanical stimulation, and the state of desensitization can last for several days. The role of histone modifications in memory gene expression and modulating plant response to abiotic or biotic signals is well known. However, such information is still lacking to explain the attenuated expression pattern of mechano-responsive genes in plants under repetitive stimulation. Using poplar as a model plant in this study, we first measured the global level of H3K9/14ac and H3K4me3 marks in the bent stem. The result shows that a single mild bending of the stem for 6 seconds is sufficient to alter the global level of the H3K9/14ac mark in poplar, highlighting the fact that plants are extremely sensitive to mechanical signals. Next, we analyzed the temporal dynamics of these two active histone marks at attenuated (PtaZFP2, PtaXET6, and PtaACA13) and non-attenuated (PtaHRD) mechano-responsive loci during the desensitization and resensitization phases. Enrichment of H3K9/14ac and H3K4me3 in the regulatory region of attenuated genes correlates well with their transient expression pattern after the first bending. Moreover, the levels of H3K4me3 correlate well with their expression pattern after the second bending at desensitization (3 days after the first bending) as well as resensitization (5 days after the first bending) phases. On the other hand, H3K9/14ac status correlates only with their attenuated expression pattern at the desensitization phase. The expression efficiency of the attenuated genes was restored after the second bending in the histone deacetylase inhibitor-treated plants. While both histone modifications contribute to the expression of attenuated genes, mechanostimulated expression of the non-attenuated PtaHRD gene seems to be H3K4me3 dependent.</p>',
'date' => '2023-02-01',
'pmid' => 'https://doi.org/10.1101%2F2023.02.12.526104',
'doi' => '10.1101/2023.02.12.526104',
'modified' => '2023-04-14 09:20:38',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4474',
'name' => 'DNA sequence and chromatin modifiers cooperate to confer epigeneticbistability at imprinting control regions.',
'authors' => 'Butz S. et al.',
'description' => '<p>Genomic imprinting is regulated by parental-specific DNA methylation of imprinting control regions (ICRs). Despite an identical DNA sequence, ICRs can exist in two distinct epigenetic states that are memorized throughout unlimited cell divisions and reset during germline formation. Here, we systematically study the genetic and epigenetic determinants of this epigenetic bistability. By iterative integration of ICRs and related DNA sequences to an ectopic location in the mouse genome, we first identify the DNA sequence features required for maintenance of epigenetic states in embryonic stem cells. The autonomous regulatory properties of ICRs further enabled us to create DNA-methylation-sensitive reporters and to screen for key components involved in regulating their epigenetic memory. Besides DNMT1, UHRF1 and ZFP57, we identify factors that prevent switching from methylated to unmethylated states and show that two of these candidates, ATF7IP and ZMYM2, are important for the stability of DNA and H3K9 methylation at ICRs in embryonic stem cells.</p>',
'date' => '2022-11-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36333500',
'doi' => '10.1038/s41588-022-01210-z',
'modified' => '2022-11-18 12:20:16',
'created' => '2022-11-15 09:26:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4345',
'name' => 'Altered Chromatin States Drive Cryptic Transcription in AgingMammalian Stem Cells.',
'authors' => 'McCauley Brenna S et al.',
'description' => '<p>A repressive chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation suppresses cryptic transcription in embryonic stem cells. Cryptic transcription is elevated with age in yeast and nematodes, and reducing it extends yeast lifespan, though whether this occurs in mammals is unknown. We show that cryptic transcription is elevated in aged mammalian stem cells, including murine hematopoietic stem cells (mHSCs) and neural stem cells (NSCs) and human mesenchymal stem cells (hMSCs). Precise mapping allowed quantification of age-associated cryptic transcription in hMSCs aged . Regions with significant age-associated cryptic transcription have a unique chromatin signature: decreased H3K36me3 and increased H3K4me1, H3K4me3, and H3K27ac with age. Genomic regions undergoing such changes resemble known promoter sequences and are bound by TBP even in young cells. Hence, the more permissive chromatin state at intragenic cryptic promoters likely underlies increased cryptic transcription in aged mammalian stem cells.</p>',
'date' => '2021-08-01',
'pmid' => 'https://doi.org/10.1038%2Fs43587-021-00091-x',
'doi' => '10.1038/s43587-021-00091-x',
'modified' => '2022-06-22 12:30:19',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '4384',
'name' => 'Age-associated cryptic transcription in mammalian stem cells is linked topermissive chromatin at cryptic promoters',
'authors' => 'McCauley B. S. et al.',
'description' => '<p>Suppressing spurious cryptic transcription by a repressive intragenic chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation is critical for maintaining self-renewal capacity in mouse embryonic stem cells. In yeast and nematodes, such cryptic transcription is elevated with age, and reducing the levels of age-associated cryptic transcription extends yeast lifespan. Whether cryptic transcription is also increased during mammalian aging is unknown. We show for the first time an age-associated elevation in cryptic transcription in several stem cell populations, including murine hematopoietic stem cells (mHSCs) and neural stem cells (NSCs) and human mesenchymal stem cells (hMSCs). Using DECAP-seq, we mapped and quantified age-associated cryptic transcription in hMSCs aged in vitro. Regions with significant age-associated cryptic transcription have a unique chromatin signature: decreased H3K36me3 and increased H3K4me1, H3K4me3, and H3K27ac with age. Furthermore, genomic regions undergoing such age-dependent chromatin changes resemble known promoter sequences and are bound by the promoter-associated protein TBP even in young cells. Hence, the more permissive chromatin state at intragenic cryptic promoters likely underlies the increase of cryptic transcription in aged mammalian stem cells.</p>',
'date' => '2020-10-01',
'pmid' => 'https://europepmc.org/article/ppr/ppr221829',
'doi' => '10.21203/rs.3.rs-82156/v1',
'modified' => '2022-08-04 16:24:46',
'created' => '2022-08-04 14:55:36',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '3990',
'name' => 'Identification of Novel Molecular Markers of Human Th17 Cells.',
'authors' => 'Sałkowska A, Karaś K, Karwaciak I, Walczak-Drzewiecka A, Krawczyk M, Sobalska-Kwapis M, Dastych J, Ratajewski M',
'description' => '<p>Th17 cells are important players in host defense against pathogens such as , , and . Th17 cell-mediated inflammation, under certain conditions in which balance in the immune system is disrupted, is the underlying pathogenic mechanism of certain autoimmune disorders, e.g., rheumatoid arthritis, Graves' disease, multiple sclerosis, and psoriasis. In the present study, using transcriptomic profiling, we selected genes and analyzed the expression of these genes to find potential novel markers of Th17 lymphocytes. We found that (apolipoprotein D); (complement component 1, Q subcomponent-like protein 1); and (cathepsin L) are expressed at significantly higher mRNA and protein levels in Th17 cells than in the Th1, Th2, and Treg subtypes. Interestingly, these genes and the proteins they encode are well associated with the function of Th17 cells, as these cells produce inflammation, which is linked with atherosclerosis and angiogenesis. Furthermore, we found that high expression of these genes in Th17 cells is associated with the acetylation of H2BK12 within their promoters. Thus, our results provide new information regarding this cell type. Based on these results, we also hope to better identify pathological conditions of clinical significance caused by Th17 cells.</p>',
'date' => '2020-07-03',
'pmid' => 'http://www.pubmed.gov/32635226',
'doi' => '10.3390/cells9071611',
'modified' => '2020-09-01 15:05:28',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '3695',
'name' => 'The Chromatin Factor HNI9 and ELONGATED HYPOCOTYL5 Maintain ROS Homeostasis under High Nitrogen Provision.',
'authors' => 'Bellegarde F, Maghiaoui A, Boucherez J, Krouk G, Lejay L, Bach L, Gojon A, Martin A',
'description' => '<p>Reactive oxygen species (ROS) can accumulate in cells at excessive levels, leading to unbalanced redox states and to potential oxidative stress, which can have damaging effects on the molecular components of plant cells. Several environmental conditions have been described as causing an elevation of ROS production in plants. Consequently, activation of detoxification responses is necessary to maintain ROS homeostasis at physiological levels. Misregulation of detoxification systems during oxidative stress can ultimately cause growth retardation and developmental defects. Here, we demonstrate that Arabidopsis () plants grown in a high nitrogen (N) environment express a set of genes involved in detoxification of ROS that maintain ROS at physiological levels. We show that the chromatin factor HIGH NITROGEN INSENSITIVE9 (HNI9) is an important mediator of this response and is required for the expression of detoxification genes. Mutation in HNI9 leads to elevated ROS levels and ROS-dependent phenotypic defects under high but not low N provision. In addition, we identify ELONGATED HYPOCOTYL5 as a major transcription factor required for activation of the detoxification program under high N. Our results demonstrate the requirement of a balance between N metabolism and ROS production, and our work establishes major regulators required to control ROS homeostasis under conditions of excess N.</p>',
'date' => '2019-05-01',
'pmid' => 'http://www.pubmed.gov/30824566',
'doi' => '10.1104/pp.18.01473',
'modified' => '2019-06-28 13:46:23',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '3579',
'name' => 'Polycomb Repressive Complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1.',
'authors' => 'Bellegarde F, Herbert L, Séré D, Caillieux E, Boucherez J, Fizames C, Roudier F, Gojon A, Martin A',
'description' => '<p>PRC2 is a major regulator of gene expression in eukaryotes. It catalyzes the repressive chromatin mark H3K27me3, which leads to very low expression of target genes. NRT2.1, which encodes a key root nitrate transporter in Arabidopsis, is targeted by H3K27me3, but the function of PRC2 on NRT2.1 remains unclear. Here, we demonstrate that PRC2 directly targets and down-regulates NRT2.1, but in a context of very high transcription, in nutritional conditions where this gene is one of the most highly expressed genes in the transcriptome. Indeed, the mutation of CLF, which encodes a PRC2 subunit, leads to a loss of H3K27me3 at NRT2.1 and results, exclusively under permissive conditions for NRT2.1, in a further increase in NRT2.1 expression, and specifically in tissues where NRT2.1 is normally expressed. Therefore, our data indicates that PRC2 tempers the hyperactivity of NRT2.1 in a context of very strong transcription. This reveals an original function of PRC2 in the control of the expression of a highly expressed gene in Arabidopsis.</p>',
'date' => '2018-05-21',
'pmid' => 'http://www.pubmed.gov/29784958',
'doi' => '10.1038/s41598-018-26349-w',
'modified' => '2019-04-17 15:55:09',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '3377',
'name' => 'Interplay of cell–cell contacts and RhoA/MRTF‐A signaling regulates cardiomyocyte identity',
'authors' => 'Dorn et al',
'description' => '<p><span>Cell–cell and cell–matrix interactions guide organ development and homeostasis by controlling lineage specification and maintenance, but the underlying molecular principles are largely unknown. Here, we show that in human developing cardiomyocytes cell–cell contacts at the intercalated disk connect to remodeling of the actin cytoskeleton by regulating the RhoA‐</span><span class="styled-content fixed-case">ROCK</span><span><span> </span>signaling to maintain an active<span> </span></span><span class="styled-content fixed-case">MRTF</span><span>/</span><span class="styled-content fixed-case">SRF</span><span><span> </span>transcriptional program essential for cardiomyocyte identity. Genetic perturbation of this mechanosensory pathway activates an ectopic fat gene program during cardiomyocyte differentiation, which ultimately primes the cells to switch to the brown/beige adipocyte lineage in response to adipogenesis‐inducing signals. We also demonstrate by<span> </span></span><em>in vivo</em><span><span> </span>fate mapping and clonal analysis of cardiac progenitors that cardiac fat and a subset of cardiac muscle arise from a common precursor expressing Isl1 and Wt1 during heart development, suggesting related mechanisms of determination between the two lineages.</span></p>',
'date' => '2018-05-15',
'pmid' => 'http://emboj.embopress.org/content/early/2018/05/15/embj.201798133',
'doi' => '10.15252/embj.201798133',
'modified' => '2018-05-22 22:50:53',
'created' => '2018-05-22 22:50:53',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '3473',
'name' => 'Neonatal exposure to hyperoxia leads to persistent disturbances in pulmonary histone signatures associated with NOS3 and STAT3 in a mouse model.',
'authors' => 'Chao CM, van den Bruck R, Lork S, Merkle J, Krampen L, Weil PP, Aydin M, Bellusci S, Jenke AC, Postberg J',
'description' => '<p>Background: Early pulmonary oxygen exposure is one of the most important factors implicated in the development of bronchopulmonary dysplasia (BPD). Methods: Here, we analyzed short- and long-term effects of neonatal hyperoxia on NOS3 and STAT3 expression and corresponding epigenetic signatures using a hyperoxia-based mouse model of BPD. Results: Early hyperoxia exposure led to a significant increase in NOS3 (median fold change × 2.37, IQR 1.54-3.68) and STAT3 (median fold change × 2.83, IQR 2.21-3.88) mRNA levels in pulmonary endothelial cells with corresponding changes in histone modification patterns such as H2aZac and H3K9ac hyperacetylation at the respective gene loci. No complete restoration in histone signatures at these loci was observed, and responsivity to later hyperoxia was altered in mouse lungs. In vitro, histone signatures in human aortic endothelial cells (HAEC) remained altered for several weeks after an initial long-term exposure to trichostatin A. This was associated with a substantial increase in baseline eNOS (median 27.2, IQR 22.3-35.6) and STAT3α (median 5.8, IQR 4.8-7.3) mRNA levels with a subsequent significant reduction in eNOS expression upon exposure to hypoxia. Conclusions: Early hyperoxia induced permanent changes in histones signatures at the NOS3 and STAT3 gene locus might partly explain the altered vascular response patterns in children with BPD.</p>',
'date' => '2018-01-01',
'pmid' => 'http://www.pubmed.gov/29581793',
'doi' => '10.1186/s13148-018-0469-0',
'modified' => '2019-02-15 21:45:12',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '3299',
'name' => 'Rapid Communication: The correlation between histone modifications and expression of key genes involved in accumulation of adipose tissue in the pig.',
'authors' => 'Kociucka B. et al.',
'description' => '<p>Histone modification is a well-known epigenetic mechanism involved in regulation of gene expression; however, it has been poorly studied in adipose tissues of the pig. Understanding the molecular background of adipose tissue development and function is essential for improving production efficiency and meat quality. The objective of this study was to identify the association between histone modification and the transcript level of genes important for lipid droplet formation and metabolism. Histone modifications at the promoter regions of 6 genes (, , , , , and ) were analyzed using a chromatin immunoprecipitation assay. Two modifications involved in activation of gene expression (acetylation of H3 histone at lysine 9 and methylation of H3 histone at lysine 4) as well as methylation of H3 histone at lysine 27, which is known to be related to gene repression, were examined. The level of histone modification was compared with transcript abundance determined using real-time PCR in tissue samples (subcutaneous fat, visceral fat, and longissimus dorsi muscle) derived from 3 pig breeds significantly differing in fatness traits (Polish Large White, Duroc, and Pietrain). Transcript levels were found to be correlated with histone modifications characteristic to active loci in 4 of 6 genes. A positive correlation between histone H3 lysine 9 acetylation modification and the transcript level of ( = 0.53, < 4.8 × 10), ( = 0.34, < 0.02), and ( = 0.43, < 1.0 × 10) genes was observed. The histone H3 lysine 4 trimethylation modification correlated with transcripts of ( = 0.64, < 4.6 × 10) and ( = 0.37, < 0.01) genes. No correlation was found between transcript level of all studied genes and histone H3 lysine 27 trimethylation level. This is the first study on histone modifications in porcine adipose tissues. We confirmed the relationship between histone modifications and expression of key genes for adipose tissue accumulation in the pig. Epigenetic modulation of the transcriptional profile of these genes (e.g., through nutritional factors) may improve porcine fatness traits in future.</p>',
'date' => '2017-10-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29108067',
'doi' => '',
'modified' => '2017-12-05 10:39:56',
'created' => '2017-12-05 09:31:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '3083',
'name' => 'Lhx2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11',
'authors' => 'Muralidharan B. et al.',
'description' => '<p>n the developing cerebral cortex, sequential transcriptional programs take neuroepithelial cells from proliferating progenitors to differentiated neurons with unique molecular identities. The regulatory changes that occur in the chromatin of the progenitors are not well understood. During deep layer neurogenesis, we show that transcription factor Lhx2 binds to distal regulatory elements of Fezf2 and Sox11, critical determinants of neuron subtype identity in the mouse neocortex. We demonstrate that Lhx2 binds to the NuRD histone remodeling complex subunits LSD1, HDAC2, and RBBP4, which are proximal regulators of the epigenetic state of chromatin. When Lhx2 is absent, active histone marks at the Fezf2 and Sox11 loci are increased. Loss of Lhx2 produces an increase, and overexpression of Lhx2 causes a decrease, in layer 5 Fezf2 and Ctip2 expressing neurons. Our results provide mechanistic insight into how Lhx2 acts as a necessary and sufficient regulator of genes that control cortical neuronal subtype identity.</p>',
'date' => '2016-12-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27909100',
'doi' => '',
'modified' => '2016-12-20 10:28:02',
'created' => '2016-12-20 10:28:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '2910',
'name' => 'Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape',
'authors' => 'Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">Previously, a role was demonstrated for transcription in the acquisition of DNA methylation at imprinted control regions in oocytes. Definition of the oocyte DNA methylome by whole genome approaches revealed that the majority of methylated CpG islands are intragenic and gene bodies are hypermethylated. Yet, the mechanisms by which transcription regulates DNA methylation in oocytes remain unclear. Here, we systematically test the link between transcription and the methylome.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We perform deep RNA-Seq and de novo transcriptome assembly at different stages of mouse oogenesis. This reveals thousands of novel non-annotated genes, as well as alternative promoters, for approximately 10 % of reference genes expressed in oocytes. In addition, a large fraction of novel promoters coincide with MaLR and ERVK transposable elements. Integration with our transcriptome assembly reveals that transcription correlates accurately with DNA methylation and accounts for approximately 85-90 % of the methylome. We generate a mouse model in which transcription across the Zac1/Plagl1 locus is abrogated in oocytes, resulting in failure of DNA methylation establishment at all CpGs of this locus. ChIP analysis in oocytes reveals H3K4me2 enrichment at the Zac1 imprinted control region when transcription is ablated, establishing a connection between transcription and chromatin remodeling at CpG islands by histone demethylases.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">By precisely defining the mouse oocyte transcriptome, this work not only highlights transcription as a cornerstone of DNA methylation establishment in female germ cells, but also provides an important resource for developmental biology research.</abstracttext></p>
</div>',
'date' => '2015-09-25',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26408185',
'doi' => '10.1186/s13059-015-0769-z',
'modified' => '2016-05-10 22:01:49',
'created' => '2016-05-10 22:01:49',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '2613',
'name' => 'CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.',
'authors' => 'Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC',
'description' => 'BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the underlying epigenetic signatures involved in its regulation. RESULTS: NOS3 and STAT3 mRNA levels were elevated in HUAEC of patients who suffered from placental insufficiency. 5-hydroxymethylcytosine, H3K9ac and Histone 2A (H2A).Zac at the NOS3 transcription start site directly correlated with NOS3 mRNA levels. Concomitantly, we observed entangled histone acetylation patterns and NOS3 response upon hypoxic conditions in vitro. Knock-down of either NOS3 or STAT3 by RNAi provided evidence for a functional NOS3/STAT3 relationship. Moreover, we recognized massive turnover of Stat3 at a discrete binding site in the NOS3 promoter. Interestingly, induced hyperacetylation resulted in short-termed increase of NOS3 mRNA followed by deferred decrease indicating that NOS3 expression could become self-attenuated by co-expressed intronic 27 nt-ncRNA. Reporter assay results and phylogenetic analyses enabled us to propose a novel model for STAT3-3'-UTR targeting by this 27-nt-ncRNA. CONCLUSIONS: An adverse intrauterine environment leads to adaptive changes of NOS3 expression. Apparently, a rapid NOS3 self-limiting response upon ectopic triggers co-exists with longer termed expression changes in response to placental insufficiency involving differential epigenetic signatures. Their persistence might contribute to impaired vascular endothelial response and consequently increase the risk of cardiovascular disease later in life.',
'date' => '2015-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25699114',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '2506',
'name' => 'Paclitaxel resistance increases oncolytic adenovirus efficacy via upregulated CAR expression and dysfunctional cell cycle control.',
'authors' => 'Ingemarsdotter CK, Tookman LA, Browne A, Pirlo K, Cutts R, Chelela C, Khurrum KF, Leung EY, Dowson S, Webber L, Khan I, Ennis D, Syed N, Crook TR, Brenton JD, Lockley M, McNeish IA',
'description' => 'Resistance to paclitaxel chemotherapy frequently develops in ovarian cancer. Oncolytic adenoviruses are a novel therapy for human malignancies that are being evaluated in early phase trials. However, there are no reliable predictive biomarkers for oncolytic adenovirus activity in ovarian cancer. We investigated the link between paclitaxel resistance and oncolytic adenovirus activity using established ovarian cancer cell line models, xenografts with de novo paclitaxel resistance and tumour samples from two separate trials. The activity of multiple Ad5 vectors, including dl922-947 (E1A CR2-deleted), dl1520 (E1B-55K deleted) and Ad5 WT, was significantly increased in paclitaxel resistant ovarian cancer in vitro and in vivo. This was associated with greater infectivity resulting from increased expression of the primary receptor for Ad5, CAR (coxsackie adenovirus receptor). This, in turn, resulted from increased CAR transcription secondary to histone modification in resistant cells. There was increased CAR expression in intraperitoneal tumours with de novo paclitaxel resistance and in tumours from patients with clinical resistance to paclitaxel. Increased CAR expression did not cause paclitaxel resistance, but did increase inflammatory cytokine expression. Finally, we identified dysregulated cell cycle control as a second mechanism of increased adenovirus efficacy in paclitaxel-resistant ovarian cancer. Ad11 and Ad35, both group B adenoviruses that utilise non-CAR receptors to infect cells, are also significantly more effective in paclitaxel-resistant ovarian cell models. Inhibition of CDK4/6 using PD-0332991 was able both to reverse paclitaxel resistance and reduce adenovirus efficacy. Thus, paclitaxel resistance increases oncolytic adenovirus efficacy via at least two separate mechanisms - if validated further, this information could have future clinical utility to aid patient selection for clinical trials.',
'date' => '2014-12-29',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25560085',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '998',
'name' => 'p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6.',
'authors' => 'Phalke S, Mzoughi S, Bezzi M, Jennifer N, Mok WC, Low DH, Thike AA, Kuznetsov VA, Tan PH, Voorhoeve PM, Guccione E',
'description' => 'p21 is a potent cyclin-dependent kinase inhibitor that plays a role in promoting G1 cell cycle arrest and cellular senescence. Consistent with this role, p21 is a downstream target of several tumour suppressors and oncogenes, and it is downregulated in the majority of tumours, including breast cancer. Here, we report that protein arginine methyltransferase 6 (PRMT6), a type I PRMT known to act as a transcriptional cofactor, directly represses the p21 promoter. PRMT6 knock-down (KD) results in a p21 derepression in breast cancer cells, which is p53-independent, and leads to cell cycle arrest, cellular senescence and reduced growth in soft agar assays and in severe combined immunodeficiency (SCID) mice for all the cancer lines examined. We finally show that bypassing the p21-mediated arrest rescues PRMT6 KD cells from senescence, and it restores their ability to grow on soft agar. We conclude that PRMT6 acts as an oncogene in breast cancer cells, promoting growth and preventing senescence, making it an attractive target for cancer therapy.',
'date' => '2012-09-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22987071',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '359',
'name' => 'Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.',
'authors' => 'Henckel A, Chebli K, Kota SK, Arnaud P, Feil R',
'description' => 'Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at 'maternal' ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.',
'date' => '2012-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22117218',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '432',
'name' => 'H3K4me3 antibody SDS US en',
'language' => 'en',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-US-en-GHS_2_0.pdf',
'countries' => 'US',
'modified' => '2020-06-30 18:23:32',
'created' => '2020-06-30 18:23:32',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '430',
'name' => 'H3K4me3 antibody SDS GB en',
'language' => 'en',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-GB-en-GHS_2_0.pdf',
'countries' => 'GB',
'modified' => '2020-06-30 18:22:36',
'created' => '2020-06-30 18:22:36',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '425',
'name' => 'H3K4me3 antibody SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-fr-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:17:31',
'created' => '2020-06-30 18:17:31',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '429',
'name' => 'H3K4me3 antibody SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-30 18:22:02',
'created' => '2020-06-30 18:22:02',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '428',
'name' => 'H3K4me3 antibody SDS ES es',
'language' => 'es',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-ES-es-GHS_2_0.pdf',
'countries' => 'ES',
'modified' => '2020-06-30 18:21:17',
'created' => '2020-06-30 18:21:17',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '427',
'name' => 'H3K4me3 antibody SDS DE de',
'language' => 'de',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-DE-de-GHS_2_0.pdf',
'countries' => 'DE',
'modified' => '2020-06-30 18:19:09',
'created' => '2020-06-30 18:19:09',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '431',
'name' => 'H3K4me3 antibody SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-JP-ja-GHS_2_0.pdf',
'countries' => 'JP',
'modified' => '2020-06-30 18:23:02',
'created' => '2020-06-30 18:23:02',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '426',
'name' => 'H3K4me3 antibody SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:18:22',
'created' => '2020-06-30 18:18:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/h3k4me3-polyclonal-antibody-classic-50-ug-25-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
)
$pro = array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/bioruptor-pico-sonication-device"><img src="/img/product/shearing_technologies/bioruptor_pico.jpg" alt="Bioruptor pico next gen sequencing " class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">B01060010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Bioruptor® Pico sonication device</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '1101',
'product_id' => '2195',
'related_id' => '1787'
),
'Image' => array(
(int) 0 => array(
'id' => '133',
'name' => 'product/shearing_technologies/bioruptor_pico.jpg',
'alt' => 'Bioruptor pico next gen sequencing ',
'modified' => '2020-11-27 10:00:30',
'created' => '2015-06-10 17:28:55',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1772',
'name' => 'product/shearing_technologies/B010600010.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 15:41:46',
'created' => '2018-02-14 15:41:46',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1773',
'name' => 'product/shearing_technologies/B010600010-2.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 16:18:03',
'created' => '2018-02-14 15:42:00',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1774',
'name' => 'product/shearing_technologies/B01060010-bioruptor-pico-cooler.jpg',
'alt' => 'Pico + cooler',
'modified' => '2018-03-06 11:01:33',
'created' => '2018-03-06 11:01:33',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'ProductsApplication' => array(
'id' => '4314',
'product_id' => '2195',
'application_id' => '42'
)
)
$slugs = array(
(int) 0 => 'chip-seq-antibodies'
)
$applications = array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'locale' => 'eng'
)
$description = ''
$name = 'ChIP-seq (ab)'
$document = array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '1616',
'product_id' => '2195',
'document_id' => '38'
)
)
$sds = array(
'id' => '426',
'name' => 'H3K4me3 antibody SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:18:22',
'created' => '2020-06-30 18:18:22',
'ProductsSafetySheet' => array(
'id' => '833',
'product_id' => '2195',
'safety_sheet_id' => '426'
)
)
$publication = array(
'id' => '359',
'name' => 'Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.',
'authors' => 'Henckel A, Chebli K, Kota SK, Arnaud P, Feil R',
'description' => 'Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at 'maternal' ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.',
'date' => '2012-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22117218',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
'id' => '870',
'product_id' => '2195',
'publication_id' => '359'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/22117218" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: header [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'product' => array(
'Product' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '114',
'name' => 'H3K4me3 polyclonal antibody',
'description' => 'Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.',
'clonality' => '',
'isotype' => '',
'lot' => '002',
'concentration' => '1.4 µg/µl',
'reactivity' => 'Human, mouse, Arabidopsis: positive. Other species: not tested.',
'type' => 'Polyclonal',
'purity' => 'Affinity purified polyclonal antibody.',
'classification' => 'Classic',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>ChIP/ChIP-seq <sup>*</sup></td>
<td>1 µg/ChIP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting</td>
<td>1:2,000</td>
<td>Fig 3</td>
</tr>
<tr>
<td>Western Blotting</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
<tr>
<td>Immunofluorescence</td>
<td>1:100</td>
<td>Fig 5</td>
</tr>
</tbody>
</table>
<p></p>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 µg per IP.</small></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide and 0.05% ProClin 300.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-11-25 17:04:17',
'created' => '0000-00-00 00:00:00',
'select_label' => '114 - H3K4me3 polyclonal antibody (002 - 1.4 µg/µl - Human, mouse, Arabidopsis: positive. Other species: not tested. - Affinity purified polyclonal antibody. - Rabbit)'
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/h3k4me3-polyclonal-antibody-classic-50-ug-25-ul'
)
$language = 'en'
$meta_keywords = 'H3K4me3 Antibody '
$meta_description = 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.'
$meta_title = 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode'
$product = array(
'Product' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '114',
'name' => 'H3K4me3 polyclonal antibody',
'description' => 'Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.',
'clonality' => '',
'isotype' => '',
'lot' => '002',
'concentration' => '1.4 µg/µl',
'reactivity' => 'Human, mouse, Arabidopsis: positive. Other species: not tested.',
'type' => 'Polyclonal',
'purity' => 'Affinity purified polyclonal antibody.',
'classification' => 'Classic',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>ChIP/ChIP-seq <sup>*</sup></td>
<td>1 µg/ChIP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting</td>
<td>1:2,000</td>
<td>Fig 3</td>
</tr>
<tr>
<td>Western Blotting</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
<tr>
<td>Immunofluorescence</td>
<td>1:100</td>
<td>Fig 5</td>
</tr>
</tbody>
</table>
<p></p>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 µg per IP.</small></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide and 0.05% ProClin 300.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-11-25 17:04:17',
'created' => '0000-00-00 00:00:00',
'select_label' => '114 - H3K4me3 polyclonal antibody (002 - 1.4 µg/µl - Human, mouse, Arabidopsis: positive. Other species: not tested. - Affinity purified polyclonal antibody. - Rabbit)'
),
'Slave' => array(
(int) 0 => array(
'id' => '322',
'name' => 'C15410030',
'product_id' => '2195',
'modified' => '2020-11-25 16:06:48',
'created' => '2020-11-25 16:06:48'
)
),
'Group' => array(
'Group' => array(
'id' => '322',
'name' => 'C15410030',
'product_id' => '2195',
'modified' => '2020-11-25 16:06:48',
'created' => '2020-11-25 16:06:48'
),
'Master' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '28',
'position' => '10',
'parent_id' => '40',
'name' => 'DB',
'description' => '<p>Dot blotting</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'dot-blotting',
'meta_keywords' => 'Dot blotting,Monoclonal & Polyclonal antibody,',
'meta_description' => 'Diagenode offers Monoclonal & Polyclonal antibodies for Dot blotting applications',
'meta_title' => 'Dot blotting Antibodies - Monoclonal & Polyclonal antibody | Diagenode',
'modified' => '2016-01-13 14:40:49',
'created' => '2015-07-08 13:45:05',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '19',
'position' => '10',
'parent_id' => '40',
'name' => 'WB',
'description' => '<p><strong>Western blot</strong> : The quality of antibodies used in this technique is crucial for correct and specific protein identification. Diagenode offers huge selection of highly sensitive and specific western blot-validated antibodies.</p>
<p>Learn more about: <a href="https://www.diagenode.com/applications/western-blot">Loading control, MW marker visualization</a><em>. <br /></em></p>
<p><em></em>Check our selection of antibodies validated in Western blot.</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'western-blot-antibodies',
'meta_keywords' => ' Western Blot Antibodies ,western blot protocol,Western Blotting Products,Polyclonal antibodies ,monoclonal antibodies ',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for western blot applications',
'meta_title' => ' Western Blot - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-26 12:44:51',
'created' => '2015-01-07 09:20:00',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '111',
'position' => '40',
'parent_id' => '4',
'name' => 'Histone antibodies',
'description' => '<p>Histones are the main protein components of chromatin involved in the compaction of DNA into nucleosomes, the basic units of chromatin. A <strong>nucleosome</strong> consists of one pair of each of the core histones (<strong>H2A</strong>, <strong>H2B</strong>, <strong>H3</strong> and <strong>H4</strong>) forming an octameric structure wrapped by 146 base pairs of DNA. The different nucleosomes are linked by the linker histone<strong> H1, </strong>allowing for further condensation of chromatin.</p>
<p>The core histones have a globular structure with large unstructured N-terminal tails protruding from the nucleosome. They can undergo to multiple post-translational modifications (PTM), mainly at the N-terminal tails. These <strong>post-translational modifications </strong>include methylation, acetylation, phosphorylation, ubiquitinylation, citrullination, sumoylation, deamination and crotonylation. The most well characterized PTMs are <strong>methylation,</strong> <strong>acetylation and phosphorylation</strong>. Histone methylation occurs mainly on lysine (K) residues, which can be mono-, di- or tri-methylated, and on arginines (R), which can be mono-methylated and symmetrically or asymmetrically di-methylated. Histone acetylation occurs on lysines and histone phosphorylation mainly on serines (S), threonines (T) and tyrosines (Y).</p>
<p>The PTMs of the different residues are involved in numerous processes such as DNA repair, DNA replication and chromosome condensation. They influence the chromatin organization and can be positively or negatively associated with gene expression. Trimethylation of H3K4, H3K36 and H3K79, and lysine acetylation generally result in an open chromatin configuration (figure below) and are therefore associated with <strong>euchromatin</strong> and gene activation. Trimethylation of H3K9, K3K27 and H4K20, on the other hand, is enriched in <strong>heterochromatin </strong>and associated with gene silencing. The combination of different histone modifications is called the "<strong>histone code</strong>”, analogous to the genetic code.</p>
<p><img src="https://www.diagenode.com/img/categories/antibodies/histone-marks-illustration.png" /></p>
<p>Diagenode is proud to offer a large range of antibodies against histones and histone modifications. Our antibodies are highly specific and have been validated in many applications, including <strong>ChIP</strong> and <strong>ChIP-seq</strong>.</p>
<p>Diagenode’s collection includes antibodies recognizing:</p>
<ul>
<li><strong>Histone H1 variants</strong></li>
<li><strong>Histone H2A, H2A variants and histone H2A</strong> <strong>modifications</strong> (serine phosphorylation, lysine acetylation, lysine ubiquitinylation)</li>
<li><strong>Histone H2B and H2B</strong> <strong>modifications </strong>(serine phosphorylation, lysine acetylation)</li>
<li><strong>Histone H3 and H3 modifications </strong>(lysine methylation (mono-, di- and tri-methylated), lysine acetylation, serine phosphorylation, threonine phosphorylation, arginine methylation (mono-methylated, symmetrically and asymmetrically di-methylated))</li>
<li><strong>Histone H4 and H4 modifications (</strong>lysine methylation (mono-, di- and tri-methylated), lysine acetylation, arginine methylation (mono-methylated and symmetrically di-methylated), serine phosphorylation )</li>
</ul>
<p><span style="font-weight: 400;"><strong>HDAC's HAT's, HMT's and other</strong> <strong>enzymes</strong> which modify histones can be found in the category <a href="../categories/chromatin-modifying-proteins-histone-transferase">Histone modifying enzymes</a><br /></span></p>
<p><span style="font-weight: 400;"> Diagenode’s highly validated antibodies:</span></p>
<ul>
<li><span style="font-weight: 400;"> Highly sensitive and specific</span></li>
<li><span style="font-weight: 400;"> Cost-effective (requires less antibody per reaction)</span></li>
<li><span style="font-weight: 400;"> Batch-specific data is available on the website</span></li>
<li><span style="font-weight: 400;"> Expert technical support</span></li>
<li><span style="font-weight: 400;"> Sample sizes available</span></li>
<li><span style="font-weight: 400;"> 100% satisfaction guarantee</span></li>
</ul>',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'histone-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Histone, antibody, histone h1, histone h2, histone h3, histone h4',
'meta_description' => 'Polyclonal and Monoclonal Antibodies against Histones and their modifications validated for many applications, including Chromatin Immunoprecipitation (ChIP) and ChIP-Sequencing (ChIP-seq)',
'meta_title' => 'Histone and Modified Histone Antibodies | Diagenode',
'modified' => '2020-09-17 13:34:56',
'created' => '2016-04-01 16:01:32',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '127',
'position' => '10',
'parent_id' => '4',
'name' => 'ChIP-grade antibodies',
'description' => '<div class="row">
<div class="small-10 columns"><center></center>
<p><br />Chromatin immunoprecipitation (<b>ChIP</b>) is a technique to study the associations of proteins with the specific genomic regions in intact cells. One of the most important steps of this protocol is the immunoprecipitation of targeted protein using the antibody specifically recognizing it. The quality of antibodies used in ChIP is essential for the success of the experiment. Diagenode offers extensively validated ChIP-grade antibodies, confirmed for their specificity, and high level of performance in ChIP. Each batch is validated, and batch-specific data are available on the website.</p>
<p></p>
</div>
<div class="small-2 columns"><img src="https://www.diagenode.com/emailing/images/epi-success-guaranteed-icon.png" alt="Epigenetic success guaranteed" /></div>
</div>
<p><strong>ChIP results</strong> obtained with the antibody directed against H3K4me3 (Cat. No. <a href="../p/h3k4me3-polyclonal-antibody-premium-50-ug-50-ul">C15410003</a>). </p>
<div class="row">
<div class="small-12 medium-6 large-6 columns"><img src="https://www.diagenode.com/img/product/antibodies/C15410003-fig1-ChIP.jpg" alt="" width="400" height="315" /> </div>
<div class="small-12 medium-6 large-6 columns">
<p></p>
<p></p>
<p></p>
</div>
</div>
<p></p>
<p>Our aim at Diagenode is to offer the largest collection of highly specific <strong>ChIP-grade antibodies</strong>. We add new antibodies monthly. Find your ChIP-grade antibody in the list below and check more information about tested applications, extensive validation data, and product information.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'chip-grade-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'ChIP-grade antibodies, polyclonal antibody, monoclonal antibody, Diagenode',
'meta_description' => 'Diagenode Offers Extensively Validated ChIP-Grade Antibodies, Confirmed for their Specificity, and high level of Performance in Chromatin Immunoprecipitation ChIP',
'meta_title' => 'Chromatin immunoprecipitation ChIP-grade antibodies | Diagenode',
'modified' => '2021-07-01 10:22:38',
'created' => '2017-02-14 11:16:04',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '405',
'name' => 'Datasheet H3K4me3 C15410030',
'description' => '<p>Datasheet description</p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_H3K4me3_C15410030.pdf',
'slug' => 'datasheet-h3k4me3-C15410030',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-11-20 17:40:11',
'created' => '2015-07-07 11:47:44',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1783',
'name' => 'product/antibodies/chipseq-grade-ab-icon.png',
'alt' => 'ChIP-seq Grade',
'modified' => '2020-11-27 07:04:40',
'created' => '2018-03-15 15:54:09',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4598',
'name' => 'SUMO protease FUG1, histone reader AL3 and the PRC1 Complex areintegral to repeat-expansion induced epigenetic silencing in Arabidopsisthaliana',
'authors' => 'Sureshkumar S. et al.',
'description' => '<p>Epigenetic gene silencing induced by expanded repeats can cause diverse phenotypes ranging from severe growth defects in plants to genetic diseases such as Friedreich’s ataxia in humans1. The molecular mechanisms underlying repeat expansion-induced epigenetic silencing remain largely unknown2,3. Using a plant model, we have previously shown that expanded repeats can induce smallRNAs which in turn can lead to epigenetic silencing through the RNA-dependent DNA methylation pathway4,5. Here, using a genetic suppressor screen, we confirm a key role for the RdDM pathway and identify novel components required for epigenetic silencing caused by expanded repeats. We show that FOURTH ULP LIKE GENE CLASS 1 (FUG1) – a SUMO protease, ALFIN-LIKE 3 – a histone reader and LIKE HETEROCHROMATIN 1 (LHP1) - a component of the PRC1 complex are required for repeat expansion-induced epigenetic silencing. Loss of any of these components suppress repeat expansion-associated phenotypes. SUMO protease FUG1 physically interacts with AL3 and perturbing its potential SUMOylation site disrupts its nuclear localisation. AL3 physically interacts with LHP1 of the PRC1 complex and the FUG1-AL3-LHP1 module is essential to confer repeat expansion-associated epigenetic silencing. Our findings highlight the importance post-translational modifiers and histone readers in epigenetic silencing caused by repeat expansions.</p>',
'date' => '2023-12-01',
'pmid' => 'https://doi.org/10.1101%2F2023.01.13.523841',
'doi' => '10.1101/2023.01.13.523841',
'modified' => '2023-04-06 09:10:33',
'created' => '2023-02-21 09:59:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4692',
'name' => 'Temporal modification of H3K9/14ac and H3K4me3 histone marksmediates mechano-responsive gene expression during the accommodationprocess in poplar',
'authors' => 'Ghosh R. et al.',
'description' => '<p>Plants can attenuate their molecular response to repetitive mechanical stimulation as a function of their mechanical history. For instance, a single bending of stem is sufficient to attenuate the gene expression in poplar plants to the subsequent mechanical stimulation, and the state of desensitization can last for several days. The role of histone modifications in memory gene expression and modulating plant response to abiotic or biotic signals is well known. However, such information is still lacking to explain the attenuated expression pattern of mechano-responsive genes in plants under repetitive stimulation. Using poplar as a model plant in this study, we first measured the global level of H3K9/14ac and H3K4me3 marks in the bent stem. The result shows that a single mild bending of the stem for 6 seconds is sufficient to alter the global level of the H3K9/14ac mark in poplar, highlighting the fact that plants are extremely sensitive to mechanical signals. Next, we analyzed the temporal dynamics of these two active histone marks at attenuated (PtaZFP2, PtaXET6, and PtaACA13) and non-attenuated (PtaHRD) mechano-responsive loci during the desensitization and resensitization phases. Enrichment of H3K9/14ac and H3K4me3 in the regulatory region of attenuated genes correlates well with their transient expression pattern after the first bending. Moreover, the levels of H3K4me3 correlate well with their expression pattern after the second bending at desensitization (3 days after the first bending) as well as resensitization (5 days after the first bending) phases. On the other hand, H3K9/14ac status correlates only with their attenuated expression pattern at the desensitization phase. The expression efficiency of the attenuated genes was restored after the second bending in the histone deacetylase inhibitor-treated plants. While both histone modifications contribute to the expression of attenuated genes, mechanostimulated expression of the non-attenuated PtaHRD gene seems to be H3K4me3 dependent.</p>',
'date' => '2023-02-01',
'pmid' => 'https://doi.org/10.1101%2F2023.02.12.526104',
'doi' => '10.1101/2023.02.12.526104',
'modified' => '2023-04-14 09:20:38',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4474',
'name' => 'DNA sequence and chromatin modifiers cooperate to confer epigeneticbistability at imprinting control regions.',
'authors' => 'Butz S. et al.',
'description' => '<p>Genomic imprinting is regulated by parental-specific DNA methylation of imprinting control regions (ICRs). Despite an identical DNA sequence, ICRs can exist in two distinct epigenetic states that are memorized throughout unlimited cell divisions and reset during germline formation. Here, we systematically study the genetic and epigenetic determinants of this epigenetic bistability. By iterative integration of ICRs and related DNA sequences to an ectopic location in the mouse genome, we first identify the DNA sequence features required for maintenance of epigenetic states in embryonic stem cells. The autonomous regulatory properties of ICRs further enabled us to create DNA-methylation-sensitive reporters and to screen for key components involved in regulating their epigenetic memory. Besides DNMT1, UHRF1 and ZFP57, we identify factors that prevent switching from methylated to unmethylated states and show that two of these candidates, ATF7IP and ZMYM2, are important for the stability of DNA and H3K9 methylation at ICRs in embryonic stem cells.</p>',
'date' => '2022-11-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36333500',
'doi' => '10.1038/s41588-022-01210-z',
'modified' => '2022-11-18 12:20:16',
'created' => '2022-11-15 09:26:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4345',
'name' => 'Altered Chromatin States Drive Cryptic Transcription in AgingMammalian Stem Cells.',
'authors' => 'McCauley Brenna S et al.',
'description' => '<p>A repressive chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation suppresses cryptic transcription in embryonic stem cells. Cryptic transcription is elevated with age in yeast and nematodes, and reducing it extends yeast lifespan, though whether this occurs in mammals is unknown. We show that cryptic transcription is elevated in aged mammalian stem cells, including murine hematopoietic stem cells (mHSCs) and neural stem cells (NSCs) and human mesenchymal stem cells (hMSCs). Precise mapping allowed quantification of age-associated cryptic transcription in hMSCs aged . Regions with significant age-associated cryptic transcription have a unique chromatin signature: decreased H3K36me3 and increased H3K4me1, H3K4me3, and H3K27ac with age. Genomic regions undergoing such changes resemble known promoter sequences and are bound by TBP even in young cells. Hence, the more permissive chromatin state at intragenic cryptic promoters likely underlies increased cryptic transcription in aged mammalian stem cells.</p>',
'date' => '2021-08-01',
'pmid' => 'https://doi.org/10.1038%2Fs43587-021-00091-x',
'doi' => '10.1038/s43587-021-00091-x',
'modified' => '2022-06-22 12:30:19',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '4384',
'name' => 'Age-associated cryptic transcription in mammalian stem cells is linked topermissive chromatin at cryptic promoters',
'authors' => 'McCauley B. S. et al.',
'description' => '<p>Suppressing spurious cryptic transcription by a repressive intragenic chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation is critical for maintaining self-renewal capacity in mouse embryonic stem cells. In yeast and nematodes, such cryptic transcription is elevated with age, and reducing the levels of age-associated cryptic transcription extends yeast lifespan. Whether cryptic transcription is also increased during mammalian aging is unknown. We show for the first time an age-associated elevation in cryptic transcription in several stem cell populations, including murine hematopoietic stem cells (mHSCs) and neural stem cells (NSCs) and human mesenchymal stem cells (hMSCs). Using DECAP-seq, we mapped and quantified age-associated cryptic transcription in hMSCs aged in vitro. Regions with significant age-associated cryptic transcription have a unique chromatin signature: decreased H3K36me3 and increased H3K4me1, H3K4me3, and H3K27ac with age. Furthermore, genomic regions undergoing such age-dependent chromatin changes resemble known promoter sequences and are bound by the promoter-associated protein TBP even in young cells. Hence, the more permissive chromatin state at intragenic cryptic promoters likely underlies the increase of cryptic transcription in aged mammalian stem cells.</p>',
'date' => '2020-10-01',
'pmid' => 'https://europepmc.org/article/ppr/ppr221829',
'doi' => '10.21203/rs.3.rs-82156/v1',
'modified' => '2022-08-04 16:24:46',
'created' => '2022-08-04 14:55:36',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '3990',
'name' => 'Identification of Novel Molecular Markers of Human Th17 Cells.',
'authors' => 'Sałkowska A, Karaś K, Karwaciak I, Walczak-Drzewiecka A, Krawczyk M, Sobalska-Kwapis M, Dastych J, Ratajewski M',
'description' => '<p>Th17 cells are important players in host defense against pathogens such as , , and . Th17 cell-mediated inflammation, under certain conditions in which balance in the immune system is disrupted, is the underlying pathogenic mechanism of certain autoimmune disorders, e.g., rheumatoid arthritis, Graves' disease, multiple sclerosis, and psoriasis. In the present study, using transcriptomic profiling, we selected genes and analyzed the expression of these genes to find potential novel markers of Th17 lymphocytes. We found that (apolipoprotein D); (complement component 1, Q subcomponent-like protein 1); and (cathepsin L) are expressed at significantly higher mRNA and protein levels in Th17 cells than in the Th1, Th2, and Treg subtypes. Interestingly, these genes and the proteins they encode are well associated with the function of Th17 cells, as these cells produce inflammation, which is linked with atherosclerosis and angiogenesis. Furthermore, we found that high expression of these genes in Th17 cells is associated with the acetylation of H2BK12 within their promoters. Thus, our results provide new information regarding this cell type. Based on these results, we also hope to better identify pathological conditions of clinical significance caused by Th17 cells.</p>',
'date' => '2020-07-03',
'pmid' => 'http://www.pubmed.gov/32635226',
'doi' => '10.3390/cells9071611',
'modified' => '2020-09-01 15:05:28',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '3695',
'name' => 'The Chromatin Factor HNI9 and ELONGATED HYPOCOTYL5 Maintain ROS Homeostasis under High Nitrogen Provision.',
'authors' => 'Bellegarde F, Maghiaoui A, Boucherez J, Krouk G, Lejay L, Bach L, Gojon A, Martin A',
'description' => '<p>Reactive oxygen species (ROS) can accumulate in cells at excessive levels, leading to unbalanced redox states and to potential oxidative stress, which can have damaging effects on the molecular components of plant cells. Several environmental conditions have been described as causing an elevation of ROS production in plants. Consequently, activation of detoxification responses is necessary to maintain ROS homeostasis at physiological levels. Misregulation of detoxification systems during oxidative stress can ultimately cause growth retardation and developmental defects. Here, we demonstrate that Arabidopsis () plants grown in a high nitrogen (N) environment express a set of genes involved in detoxification of ROS that maintain ROS at physiological levels. We show that the chromatin factor HIGH NITROGEN INSENSITIVE9 (HNI9) is an important mediator of this response and is required for the expression of detoxification genes. Mutation in HNI9 leads to elevated ROS levels and ROS-dependent phenotypic defects under high but not low N provision. In addition, we identify ELONGATED HYPOCOTYL5 as a major transcription factor required for activation of the detoxification program under high N. Our results demonstrate the requirement of a balance between N metabolism and ROS production, and our work establishes major regulators required to control ROS homeostasis under conditions of excess N.</p>',
'date' => '2019-05-01',
'pmid' => 'http://www.pubmed.gov/30824566',
'doi' => '10.1104/pp.18.01473',
'modified' => '2019-06-28 13:46:23',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '3579',
'name' => 'Polycomb Repressive Complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1.',
'authors' => 'Bellegarde F, Herbert L, Séré D, Caillieux E, Boucherez J, Fizames C, Roudier F, Gojon A, Martin A',
'description' => '<p>PRC2 is a major regulator of gene expression in eukaryotes. It catalyzes the repressive chromatin mark H3K27me3, which leads to very low expression of target genes. NRT2.1, which encodes a key root nitrate transporter in Arabidopsis, is targeted by H3K27me3, but the function of PRC2 on NRT2.1 remains unclear. Here, we demonstrate that PRC2 directly targets and down-regulates NRT2.1, but in a context of very high transcription, in nutritional conditions where this gene is one of the most highly expressed genes in the transcriptome. Indeed, the mutation of CLF, which encodes a PRC2 subunit, leads to a loss of H3K27me3 at NRT2.1 and results, exclusively under permissive conditions for NRT2.1, in a further increase in NRT2.1 expression, and specifically in tissues where NRT2.1 is normally expressed. Therefore, our data indicates that PRC2 tempers the hyperactivity of NRT2.1 in a context of very strong transcription. This reveals an original function of PRC2 in the control of the expression of a highly expressed gene in Arabidopsis.</p>',
'date' => '2018-05-21',
'pmid' => 'http://www.pubmed.gov/29784958',
'doi' => '10.1038/s41598-018-26349-w',
'modified' => '2019-04-17 15:55:09',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '3377',
'name' => 'Interplay of cell–cell contacts and RhoA/MRTF‐A signaling regulates cardiomyocyte identity',
'authors' => 'Dorn et al',
'description' => '<p><span>Cell–cell and cell–matrix interactions guide organ development and homeostasis by controlling lineage specification and maintenance, but the underlying molecular principles are largely unknown. Here, we show that in human developing cardiomyocytes cell–cell contacts at the intercalated disk connect to remodeling of the actin cytoskeleton by regulating the RhoA‐</span><span class="styled-content fixed-case">ROCK</span><span><span> </span>signaling to maintain an active<span> </span></span><span class="styled-content fixed-case">MRTF</span><span>/</span><span class="styled-content fixed-case">SRF</span><span><span> </span>transcriptional program essential for cardiomyocyte identity. Genetic perturbation of this mechanosensory pathway activates an ectopic fat gene program during cardiomyocyte differentiation, which ultimately primes the cells to switch to the brown/beige adipocyte lineage in response to adipogenesis‐inducing signals. We also demonstrate by<span> </span></span><em>in vivo</em><span><span> </span>fate mapping and clonal analysis of cardiac progenitors that cardiac fat and a subset of cardiac muscle arise from a common precursor expressing Isl1 and Wt1 during heart development, suggesting related mechanisms of determination between the two lineages.</span></p>',
'date' => '2018-05-15',
'pmid' => 'http://emboj.embopress.org/content/early/2018/05/15/embj.201798133',
'doi' => '10.15252/embj.201798133',
'modified' => '2018-05-22 22:50:53',
'created' => '2018-05-22 22:50:53',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '3473',
'name' => 'Neonatal exposure to hyperoxia leads to persistent disturbances in pulmonary histone signatures associated with NOS3 and STAT3 in a mouse model.',
'authors' => 'Chao CM, van den Bruck R, Lork S, Merkle J, Krampen L, Weil PP, Aydin M, Bellusci S, Jenke AC, Postberg J',
'description' => '<p>Background: Early pulmonary oxygen exposure is one of the most important factors implicated in the development of bronchopulmonary dysplasia (BPD). Methods: Here, we analyzed short- and long-term effects of neonatal hyperoxia on NOS3 and STAT3 expression and corresponding epigenetic signatures using a hyperoxia-based mouse model of BPD. Results: Early hyperoxia exposure led to a significant increase in NOS3 (median fold change × 2.37, IQR 1.54-3.68) and STAT3 (median fold change × 2.83, IQR 2.21-3.88) mRNA levels in pulmonary endothelial cells with corresponding changes in histone modification patterns such as H2aZac and H3K9ac hyperacetylation at the respective gene loci. No complete restoration in histone signatures at these loci was observed, and responsivity to later hyperoxia was altered in mouse lungs. In vitro, histone signatures in human aortic endothelial cells (HAEC) remained altered for several weeks after an initial long-term exposure to trichostatin A. This was associated with a substantial increase in baseline eNOS (median 27.2, IQR 22.3-35.6) and STAT3α (median 5.8, IQR 4.8-7.3) mRNA levels with a subsequent significant reduction in eNOS expression upon exposure to hypoxia. Conclusions: Early hyperoxia induced permanent changes in histones signatures at the NOS3 and STAT3 gene locus might partly explain the altered vascular response patterns in children with BPD.</p>',
'date' => '2018-01-01',
'pmid' => 'http://www.pubmed.gov/29581793',
'doi' => '10.1186/s13148-018-0469-0',
'modified' => '2019-02-15 21:45:12',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '3299',
'name' => 'Rapid Communication: The correlation between histone modifications and expression of key genes involved in accumulation of adipose tissue in the pig.',
'authors' => 'Kociucka B. et al.',
'description' => '<p>Histone modification is a well-known epigenetic mechanism involved in regulation of gene expression; however, it has been poorly studied in adipose tissues of the pig. Understanding the molecular background of adipose tissue development and function is essential for improving production efficiency and meat quality. The objective of this study was to identify the association between histone modification and the transcript level of genes important for lipid droplet formation and metabolism. Histone modifications at the promoter regions of 6 genes (, , , , , and ) were analyzed using a chromatin immunoprecipitation assay. Two modifications involved in activation of gene expression (acetylation of H3 histone at lysine 9 and methylation of H3 histone at lysine 4) as well as methylation of H3 histone at lysine 27, which is known to be related to gene repression, were examined. The level of histone modification was compared with transcript abundance determined using real-time PCR in tissue samples (subcutaneous fat, visceral fat, and longissimus dorsi muscle) derived from 3 pig breeds significantly differing in fatness traits (Polish Large White, Duroc, and Pietrain). Transcript levels were found to be correlated with histone modifications characteristic to active loci in 4 of 6 genes. A positive correlation between histone H3 lysine 9 acetylation modification and the transcript level of ( = 0.53, < 4.8 × 10), ( = 0.34, < 0.02), and ( = 0.43, < 1.0 × 10) genes was observed. The histone H3 lysine 4 trimethylation modification correlated with transcripts of ( = 0.64, < 4.6 × 10) and ( = 0.37, < 0.01) genes. No correlation was found between transcript level of all studied genes and histone H3 lysine 27 trimethylation level. This is the first study on histone modifications in porcine adipose tissues. We confirmed the relationship between histone modifications and expression of key genes for adipose tissue accumulation in the pig. Epigenetic modulation of the transcriptional profile of these genes (e.g., through nutritional factors) may improve porcine fatness traits in future.</p>',
'date' => '2017-10-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29108067',
'doi' => '',
'modified' => '2017-12-05 10:39:56',
'created' => '2017-12-05 09:31:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '3083',
'name' => 'Lhx2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11',
'authors' => 'Muralidharan B. et al.',
'description' => '<p>n the developing cerebral cortex, sequential transcriptional programs take neuroepithelial cells from proliferating progenitors to differentiated neurons with unique molecular identities. The regulatory changes that occur in the chromatin of the progenitors are not well understood. During deep layer neurogenesis, we show that transcription factor Lhx2 binds to distal regulatory elements of Fezf2 and Sox11, critical determinants of neuron subtype identity in the mouse neocortex. We demonstrate that Lhx2 binds to the NuRD histone remodeling complex subunits LSD1, HDAC2, and RBBP4, which are proximal regulators of the epigenetic state of chromatin. When Lhx2 is absent, active histone marks at the Fezf2 and Sox11 loci are increased. Loss of Lhx2 produces an increase, and overexpression of Lhx2 causes a decrease, in layer 5 Fezf2 and Ctip2 expressing neurons. Our results provide mechanistic insight into how Lhx2 acts as a necessary and sufficient regulator of genes that control cortical neuronal subtype identity.</p>',
'date' => '2016-12-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27909100',
'doi' => '',
'modified' => '2016-12-20 10:28:02',
'created' => '2016-12-20 10:28:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '2910',
'name' => 'Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape',
'authors' => 'Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">Previously, a role was demonstrated for transcription in the acquisition of DNA methylation at imprinted control regions in oocytes. Definition of the oocyte DNA methylome by whole genome approaches revealed that the majority of methylated CpG islands are intragenic and gene bodies are hypermethylated. Yet, the mechanisms by which transcription regulates DNA methylation in oocytes remain unclear. Here, we systematically test the link between transcription and the methylome.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We perform deep RNA-Seq and de novo transcriptome assembly at different stages of mouse oogenesis. This reveals thousands of novel non-annotated genes, as well as alternative promoters, for approximately 10 % of reference genes expressed in oocytes. In addition, a large fraction of novel promoters coincide with MaLR and ERVK transposable elements. Integration with our transcriptome assembly reveals that transcription correlates accurately with DNA methylation and accounts for approximately 85-90 % of the methylome. We generate a mouse model in which transcription across the Zac1/Plagl1 locus is abrogated in oocytes, resulting in failure of DNA methylation establishment at all CpGs of this locus. ChIP analysis in oocytes reveals H3K4me2 enrichment at the Zac1 imprinted control region when transcription is ablated, establishing a connection between transcription and chromatin remodeling at CpG islands by histone demethylases.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">By precisely defining the mouse oocyte transcriptome, this work not only highlights transcription as a cornerstone of DNA methylation establishment in female germ cells, but also provides an important resource for developmental biology research.</abstracttext></p>
</div>',
'date' => '2015-09-25',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26408185',
'doi' => '10.1186/s13059-015-0769-z',
'modified' => '2016-05-10 22:01:49',
'created' => '2016-05-10 22:01:49',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '2613',
'name' => 'CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.',
'authors' => 'Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC',
'description' => 'BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the underlying epigenetic signatures involved in its regulation. RESULTS: NOS3 and STAT3 mRNA levels were elevated in HUAEC of patients who suffered from placental insufficiency. 5-hydroxymethylcytosine, H3K9ac and Histone 2A (H2A).Zac at the NOS3 transcription start site directly correlated with NOS3 mRNA levels. Concomitantly, we observed entangled histone acetylation patterns and NOS3 response upon hypoxic conditions in vitro. Knock-down of either NOS3 or STAT3 by RNAi provided evidence for a functional NOS3/STAT3 relationship. Moreover, we recognized massive turnover of Stat3 at a discrete binding site in the NOS3 promoter. Interestingly, induced hyperacetylation resulted in short-termed increase of NOS3 mRNA followed by deferred decrease indicating that NOS3 expression could become self-attenuated by co-expressed intronic 27 nt-ncRNA. Reporter assay results and phylogenetic analyses enabled us to propose a novel model for STAT3-3'-UTR targeting by this 27-nt-ncRNA. CONCLUSIONS: An adverse intrauterine environment leads to adaptive changes of NOS3 expression. Apparently, a rapid NOS3 self-limiting response upon ectopic triggers co-exists with longer termed expression changes in response to placental insufficiency involving differential epigenetic signatures. Their persistence might contribute to impaired vascular endothelial response and consequently increase the risk of cardiovascular disease later in life.',
'date' => '2015-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25699114',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '2506',
'name' => 'Paclitaxel resistance increases oncolytic adenovirus efficacy via upregulated CAR expression and dysfunctional cell cycle control.',
'authors' => 'Ingemarsdotter CK, Tookman LA, Browne A, Pirlo K, Cutts R, Chelela C, Khurrum KF, Leung EY, Dowson S, Webber L, Khan I, Ennis D, Syed N, Crook TR, Brenton JD, Lockley M, McNeish IA',
'description' => 'Resistance to paclitaxel chemotherapy frequently develops in ovarian cancer. Oncolytic adenoviruses are a novel therapy for human malignancies that are being evaluated in early phase trials. However, there are no reliable predictive biomarkers for oncolytic adenovirus activity in ovarian cancer. We investigated the link between paclitaxel resistance and oncolytic adenovirus activity using established ovarian cancer cell line models, xenografts with de novo paclitaxel resistance and tumour samples from two separate trials. The activity of multiple Ad5 vectors, including dl922-947 (E1A CR2-deleted), dl1520 (E1B-55K deleted) and Ad5 WT, was significantly increased in paclitaxel resistant ovarian cancer in vitro and in vivo. This was associated with greater infectivity resulting from increased expression of the primary receptor for Ad5, CAR (coxsackie adenovirus receptor). This, in turn, resulted from increased CAR transcription secondary to histone modification in resistant cells. There was increased CAR expression in intraperitoneal tumours with de novo paclitaxel resistance and in tumours from patients with clinical resistance to paclitaxel. Increased CAR expression did not cause paclitaxel resistance, but did increase inflammatory cytokine expression. Finally, we identified dysregulated cell cycle control as a second mechanism of increased adenovirus efficacy in paclitaxel-resistant ovarian cancer. Ad11 and Ad35, both group B adenoviruses that utilise non-CAR receptors to infect cells, are also significantly more effective in paclitaxel-resistant ovarian cell models. Inhibition of CDK4/6 using PD-0332991 was able both to reverse paclitaxel resistance and reduce adenovirus efficacy. Thus, paclitaxel resistance increases oncolytic adenovirus efficacy via at least two separate mechanisms - if validated further, this information could have future clinical utility to aid patient selection for clinical trials.',
'date' => '2014-12-29',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25560085',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '998',
'name' => 'p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6.',
'authors' => 'Phalke S, Mzoughi S, Bezzi M, Jennifer N, Mok WC, Low DH, Thike AA, Kuznetsov VA, Tan PH, Voorhoeve PM, Guccione E',
'description' => 'p21 is a potent cyclin-dependent kinase inhibitor that plays a role in promoting G1 cell cycle arrest and cellular senescence. Consistent with this role, p21 is a downstream target of several tumour suppressors and oncogenes, and it is downregulated in the majority of tumours, including breast cancer. Here, we report that protein arginine methyltransferase 6 (PRMT6), a type I PRMT known to act as a transcriptional cofactor, directly represses the p21 promoter. PRMT6 knock-down (KD) results in a p21 derepression in breast cancer cells, which is p53-independent, and leads to cell cycle arrest, cellular senescence and reduced growth in soft agar assays and in severe combined immunodeficiency (SCID) mice for all the cancer lines examined. We finally show that bypassing the p21-mediated arrest rescues PRMT6 KD cells from senescence, and it restores their ability to grow on soft agar. We conclude that PRMT6 acts as an oncogene in breast cancer cells, promoting growth and preventing senescence, making it an attractive target for cancer therapy.',
'date' => '2012-09-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22987071',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '359',
'name' => 'Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.',
'authors' => 'Henckel A, Chebli K, Kota SK, Arnaud P, Feil R',
'description' => 'Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at 'maternal' ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.',
'date' => '2012-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22117218',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '432',
'name' => 'H3K4me3 antibody SDS US en',
'language' => 'en',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-US-en-GHS_2_0.pdf',
'countries' => 'US',
'modified' => '2020-06-30 18:23:32',
'created' => '2020-06-30 18:23:32',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '430',
'name' => 'H3K4me3 antibody SDS GB en',
'language' => 'en',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-GB-en-GHS_2_0.pdf',
'countries' => 'GB',
'modified' => '2020-06-30 18:22:36',
'created' => '2020-06-30 18:22:36',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '425',
'name' => 'H3K4me3 antibody SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-fr-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:17:31',
'created' => '2020-06-30 18:17:31',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '429',
'name' => 'H3K4me3 antibody SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-30 18:22:02',
'created' => '2020-06-30 18:22:02',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '428',
'name' => 'H3K4me3 antibody SDS ES es',
'language' => 'es',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-ES-es-GHS_2_0.pdf',
'countries' => 'ES',
'modified' => '2020-06-30 18:21:17',
'created' => '2020-06-30 18:21:17',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '427',
'name' => 'H3K4me3 antibody SDS DE de',
'language' => 'de',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-DE-de-GHS_2_0.pdf',
'countries' => 'DE',
'modified' => '2020-06-30 18:19:09',
'created' => '2020-06-30 18:19:09',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '431',
'name' => 'H3K4me3 antibody SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-JP-ja-GHS_2_0.pdf',
'countries' => 'JP',
'modified' => '2020-06-30 18:23:02',
'created' => '2020-06-30 18:23:02',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '426',
'name' => 'H3K4me3 antibody SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:18:22',
'created' => '2020-06-30 18:18:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/h3k4me3-polyclonal-antibody-classic-50-ug-25-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
)
$pro = array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/bioruptor-pico-sonication-device"><img src="/img/product/shearing_technologies/bioruptor_pico.jpg" alt="Bioruptor pico next gen sequencing " class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">B01060010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Bioruptor® Pico sonication device</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '1101',
'product_id' => '2195',
'related_id' => '1787'
),
'Image' => array(
(int) 0 => array(
'id' => '133',
'name' => 'product/shearing_technologies/bioruptor_pico.jpg',
'alt' => 'Bioruptor pico next gen sequencing ',
'modified' => '2020-11-27 10:00:30',
'created' => '2015-06-10 17:28:55',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1772',
'name' => 'product/shearing_technologies/B010600010.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 15:41:46',
'created' => '2018-02-14 15:41:46',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1773',
'name' => 'product/shearing_technologies/B010600010-2.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 16:18:03',
'created' => '2018-02-14 15:42:00',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1774',
'name' => 'product/shearing_technologies/B01060010-bioruptor-pico-cooler.jpg',
'alt' => 'Pico + cooler',
'modified' => '2018-03-06 11:01:33',
'created' => '2018-03-06 11:01:33',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'ProductsApplication' => array(
'id' => '4314',
'product_id' => '2195',
'application_id' => '42'
)
)
$slugs = array(
(int) 0 => 'chip-seq-antibodies'
)
$applications = array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'locale' => 'eng'
)
$description = ''
$name = 'ChIP-seq (ab)'
$document = array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '1616',
'product_id' => '2195',
'document_id' => '38'
)
)
$sds = array(
'id' => '426',
'name' => 'H3K4me3 antibody SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:18:22',
'created' => '2020-06-30 18:18:22',
'ProductsSafetySheet' => array(
'id' => '833',
'product_id' => '2195',
'safety_sheet_id' => '426'
)
)
$publication = array(
'id' => '359',
'name' => 'Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.',
'authors' => 'Henckel A, Chebli K, Kota SK, Arnaud P, Feil R',
'description' => 'Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at 'maternal' ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.',
'date' => '2012-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22117218',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
'id' => '870',
'product_id' => '2195',
'publication_id' => '359'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/22117218" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: message [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'product' => array(
'Product' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '114',
'name' => 'H3K4me3 polyclonal antibody',
'description' => 'Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.',
'clonality' => '',
'isotype' => '',
'lot' => '002',
'concentration' => '1.4 µg/µl',
'reactivity' => 'Human, mouse, Arabidopsis: positive. Other species: not tested.',
'type' => 'Polyclonal',
'purity' => 'Affinity purified polyclonal antibody.',
'classification' => 'Classic',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>ChIP/ChIP-seq <sup>*</sup></td>
<td>1 µg/ChIP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting</td>
<td>1:2,000</td>
<td>Fig 3</td>
</tr>
<tr>
<td>Western Blotting</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
<tr>
<td>Immunofluorescence</td>
<td>1:100</td>
<td>Fig 5</td>
</tr>
</tbody>
</table>
<p></p>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 µg per IP.</small></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide and 0.05% ProClin 300.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-11-25 17:04:17',
'created' => '0000-00-00 00:00:00',
'select_label' => '114 - H3K4me3 polyclonal antibody (002 - 1.4 µg/µl - Human, mouse, Arabidopsis: positive. Other species: not tested. - Affinity purified polyclonal antibody. - Rabbit)'
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/h3k4me3-polyclonal-antibody-classic-50-ug-25-ul'
)
$language = 'en'
$meta_keywords = 'H3K4me3 Antibody '
$meta_description = 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.'
$meta_title = 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode'
$product = array(
'Product' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '114',
'name' => 'H3K4me3 polyclonal antibody',
'description' => 'Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.',
'clonality' => '',
'isotype' => '',
'lot' => '002',
'concentration' => '1.4 µg/µl',
'reactivity' => 'Human, mouse, Arabidopsis: positive. Other species: not tested.',
'type' => 'Polyclonal',
'purity' => 'Affinity purified polyclonal antibody.',
'classification' => 'Classic',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>ChIP/ChIP-seq <sup>*</sup></td>
<td>1 µg/ChIP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting</td>
<td>1:2,000</td>
<td>Fig 3</td>
</tr>
<tr>
<td>Western Blotting</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
<tr>
<td>Immunofluorescence</td>
<td>1:100</td>
<td>Fig 5</td>
</tr>
</tbody>
</table>
<p></p>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 µg per IP.</small></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide and 0.05% ProClin 300.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-11-25 17:04:17',
'created' => '0000-00-00 00:00:00',
'select_label' => '114 - H3K4me3 polyclonal antibody (002 - 1.4 µg/µl - Human, mouse, Arabidopsis: positive. Other species: not tested. - Affinity purified polyclonal antibody. - Rabbit)'
),
'Slave' => array(
(int) 0 => array(
'id' => '322',
'name' => 'C15410030',
'product_id' => '2195',
'modified' => '2020-11-25 16:06:48',
'created' => '2020-11-25 16:06:48'
)
),
'Group' => array(
'Group' => array(
'id' => '322',
'name' => 'C15410030',
'product_id' => '2195',
'modified' => '2020-11-25 16:06:48',
'created' => '2020-11-25 16:06:48'
),
'Master' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '28',
'position' => '10',
'parent_id' => '40',
'name' => 'DB',
'description' => '<p>Dot blotting</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'dot-blotting',
'meta_keywords' => 'Dot blotting,Monoclonal & Polyclonal antibody,',
'meta_description' => 'Diagenode offers Monoclonal & Polyclonal antibodies for Dot blotting applications',
'meta_title' => 'Dot blotting Antibodies - Monoclonal & Polyclonal antibody | Diagenode',
'modified' => '2016-01-13 14:40:49',
'created' => '2015-07-08 13:45:05',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '19',
'position' => '10',
'parent_id' => '40',
'name' => 'WB',
'description' => '<p><strong>Western blot</strong> : The quality of antibodies used in this technique is crucial for correct and specific protein identification. Diagenode offers huge selection of highly sensitive and specific western blot-validated antibodies.</p>
<p>Learn more about: <a href="https://www.diagenode.com/applications/western-blot">Loading control, MW marker visualization</a><em>. <br /></em></p>
<p><em></em>Check our selection of antibodies validated in Western blot.</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'western-blot-antibodies',
'meta_keywords' => ' Western Blot Antibodies ,western blot protocol,Western Blotting Products,Polyclonal antibodies ,monoclonal antibodies ',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for western blot applications',
'meta_title' => ' Western Blot - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-26 12:44:51',
'created' => '2015-01-07 09:20:00',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '111',
'position' => '40',
'parent_id' => '4',
'name' => 'Histone antibodies',
'description' => '<p>Histones are the main protein components of chromatin involved in the compaction of DNA into nucleosomes, the basic units of chromatin. A <strong>nucleosome</strong> consists of one pair of each of the core histones (<strong>H2A</strong>, <strong>H2B</strong>, <strong>H3</strong> and <strong>H4</strong>) forming an octameric structure wrapped by 146 base pairs of DNA. The different nucleosomes are linked by the linker histone<strong> H1, </strong>allowing for further condensation of chromatin.</p>
<p>The core histones have a globular structure with large unstructured N-terminal tails protruding from the nucleosome. They can undergo to multiple post-translational modifications (PTM), mainly at the N-terminal tails. These <strong>post-translational modifications </strong>include methylation, acetylation, phosphorylation, ubiquitinylation, citrullination, sumoylation, deamination and crotonylation. The most well characterized PTMs are <strong>methylation,</strong> <strong>acetylation and phosphorylation</strong>. Histone methylation occurs mainly on lysine (K) residues, which can be mono-, di- or tri-methylated, and on arginines (R), which can be mono-methylated and symmetrically or asymmetrically di-methylated. Histone acetylation occurs on lysines and histone phosphorylation mainly on serines (S), threonines (T) and tyrosines (Y).</p>
<p>The PTMs of the different residues are involved in numerous processes such as DNA repair, DNA replication and chromosome condensation. They influence the chromatin organization and can be positively or negatively associated with gene expression. Trimethylation of H3K4, H3K36 and H3K79, and lysine acetylation generally result in an open chromatin configuration (figure below) and are therefore associated with <strong>euchromatin</strong> and gene activation. Trimethylation of H3K9, K3K27 and H4K20, on the other hand, is enriched in <strong>heterochromatin </strong>and associated with gene silencing. The combination of different histone modifications is called the "<strong>histone code</strong>”, analogous to the genetic code.</p>
<p><img src="https://www.diagenode.com/img/categories/antibodies/histone-marks-illustration.png" /></p>
<p>Diagenode is proud to offer a large range of antibodies against histones and histone modifications. Our antibodies are highly specific and have been validated in many applications, including <strong>ChIP</strong> and <strong>ChIP-seq</strong>.</p>
<p>Diagenode’s collection includes antibodies recognizing:</p>
<ul>
<li><strong>Histone H1 variants</strong></li>
<li><strong>Histone H2A, H2A variants and histone H2A</strong> <strong>modifications</strong> (serine phosphorylation, lysine acetylation, lysine ubiquitinylation)</li>
<li><strong>Histone H2B and H2B</strong> <strong>modifications </strong>(serine phosphorylation, lysine acetylation)</li>
<li><strong>Histone H3 and H3 modifications </strong>(lysine methylation (mono-, di- and tri-methylated), lysine acetylation, serine phosphorylation, threonine phosphorylation, arginine methylation (mono-methylated, symmetrically and asymmetrically di-methylated))</li>
<li><strong>Histone H4 and H4 modifications (</strong>lysine methylation (mono-, di- and tri-methylated), lysine acetylation, arginine methylation (mono-methylated and symmetrically di-methylated), serine phosphorylation )</li>
</ul>
<p><span style="font-weight: 400;"><strong>HDAC's HAT's, HMT's and other</strong> <strong>enzymes</strong> which modify histones can be found in the category <a href="../categories/chromatin-modifying-proteins-histone-transferase">Histone modifying enzymes</a><br /></span></p>
<p><span style="font-weight: 400;"> Diagenode’s highly validated antibodies:</span></p>
<ul>
<li><span style="font-weight: 400;"> Highly sensitive and specific</span></li>
<li><span style="font-weight: 400;"> Cost-effective (requires less antibody per reaction)</span></li>
<li><span style="font-weight: 400;"> Batch-specific data is available on the website</span></li>
<li><span style="font-weight: 400;"> Expert technical support</span></li>
<li><span style="font-weight: 400;"> Sample sizes available</span></li>
<li><span style="font-weight: 400;"> 100% satisfaction guarantee</span></li>
</ul>',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'histone-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Histone, antibody, histone h1, histone h2, histone h3, histone h4',
'meta_description' => 'Polyclonal and Monoclonal Antibodies against Histones and their modifications validated for many applications, including Chromatin Immunoprecipitation (ChIP) and ChIP-Sequencing (ChIP-seq)',
'meta_title' => 'Histone and Modified Histone Antibodies | Diagenode',
'modified' => '2020-09-17 13:34:56',
'created' => '2016-04-01 16:01:32',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '127',
'position' => '10',
'parent_id' => '4',
'name' => 'ChIP-grade antibodies',
'description' => '<div class="row">
<div class="small-10 columns"><center></center>
<p><br />Chromatin immunoprecipitation (<b>ChIP</b>) is a technique to study the associations of proteins with the specific genomic regions in intact cells. One of the most important steps of this protocol is the immunoprecipitation of targeted protein using the antibody specifically recognizing it. The quality of antibodies used in ChIP is essential for the success of the experiment. Diagenode offers extensively validated ChIP-grade antibodies, confirmed for their specificity, and high level of performance in ChIP. Each batch is validated, and batch-specific data are available on the website.</p>
<p></p>
</div>
<div class="small-2 columns"><img src="https://www.diagenode.com/emailing/images/epi-success-guaranteed-icon.png" alt="Epigenetic success guaranteed" /></div>
</div>
<p><strong>ChIP results</strong> obtained with the antibody directed against H3K4me3 (Cat. No. <a href="../p/h3k4me3-polyclonal-antibody-premium-50-ug-50-ul">C15410003</a>). </p>
<div class="row">
<div class="small-12 medium-6 large-6 columns"><img src="https://www.diagenode.com/img/product/antibodies/C15410003-fig1-ChIP.jpg" alt="" width="400" height="315" /> </div>
<div class="small-12 medium-6 large-6 columns">
<p></p>
<p></p>
<p></p>
</div>
</div>
<p></p>
<p>Our aim at Diagenode is to offer the largest collection of highly specific <strong>ChIP-grade antibodies</strong>. We add new antibodies monthly. Find your ChIP-grade antibody in the list below and check more information about tested applications, extensive validation data, and product information.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'chip-grade-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'ChIP-grade antibodies, polyclonal antibody, monoclonal antibody, Diagenode',
'meta_description' => 'Diagenode Offers Extensively Validated ChIP-Grade Antibodies, Confirmed for their Specificity, and high level of Performance in Chromatin Immunoprecipitation ChIP',
'meta_title' => 'Chromatin immunoprecipitation ChIP-grade antibodies | Diagenode',
'modified' => '2021-07-01 10:22:38',
'created' => '2017-02-14 11:16:04',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '405',
'name' => 'Datasheet H3K4me3 C15410030',
'description' => '<p>Datasheet description</p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_H3K4me3_C15410030.pdf',
'slug' => 'datasheet-h3k4me3-C15410030',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-11-20 17:40:11',
'created' => '2015-07-07 11:47:44',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1783',
'name' => 'product/antibodies/chipseq-grade-ab-icon.png',
'alt' => 'ChIP-seq Grade',
'modified' => '2020-11-27 07:04:40',
'created' => '2018-03-15 15:54:09',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4598',
'name' => 'SUMO protease FUG1, histone reader AL3 and the PRC1 Complex areintegral to repeat-expansion induced epigenetic silencing in Arabidopsisthaliana',
'authors' => 'Sureshkumar S. et al.',
'description' => '<p>Epigenetic gene silencing induced by expanded repeats can cause diverse phenotypes ranging from severe growth defects in plants to genetic diseases such as Friedreich’s ataxia in humans1. The molecular mechanisms underlying repeat expansion-induced epigenetic silencing remain largely unknown2,3. Using a plant model, we have previously shown that expanded repeats can induce smallRNAs which in turn can lead to epigenetic silencing through the RNA-dependent DNA methylation pathway4,5. Here, using a genetic suppressor screen, we confirm a key role for the RdDM pathway and identify novel components required for epigenetic silencing caused by expanded repeats. We show that FOURTH ULP LIKE GENE CLASS 1 (FUG1) – a SUMO protease, ALFIN-LIKE 3 – a histone reader and LIKE HETEROCHROMATIN 1 (LHP1) - a component of the PRC1 complex are required for repeat expansion-induced epigenetic silencing. Loss of any of these components suppress repeat expansion-associated phenotypes. SUMO protease FUG1 physically interacts with AL3 and perturbing its potential SUMOylation site disrupts its nuclear localisation. AL3 physically interacts with LHP1 of the PRC1 complex and the FUG1-AL3-LHP1 module is essential to confer repeat expansion-associated epigenetic silencing. Our findings highlight the importance post-translational modifiers and histone readers in epigenetic silencing caused by repeat expansions.</p>',
'date' => '2023-12-01',
'pmid' => 'https://doi.org/10.1101%2F2023.01.13.523841',
'doi' => '10.1101/2023.01.13.523841',
'modified' => '2023-04-06 09:10:33',
'created' => '2023-02-21 09:59:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4692',
'name' => 'Temporal modification of H3K9/14ac and H3K4me3 histone marksmediates mechano-responsive gene expression during the accommodationprocess in poplar',
'authors' => 'Ghosh R. et al.',
'description' => '<p>Plants can attenuate their molecular response to repetitive mechanical stimulation as a function of their mechanical history. For instance, a single bending of stem is sufficient to attenuate the gene expression in poplar plants to the subsequent mechanical stimulation, and the state of desensitization can last for several days. The role of histone modifications in memory gene expression and modulating plant response to abiotic or biotic signals is well known. However, such information is still lacking to explain the attenuated expression pattern of mechano-responsive genes in plants under repetitive stimulation. Using poplar as a model plant in this study, we first measured the global level of H3K9/14ac and H3K4me3 marks in the bent stem. The result shows that a single mild bending of the stem for 6 seconds is sufficient to alter the global level of the H3K9/14ac mark in poplar, highlighting the fact that plants are extremely sensitive to mechanical signals. Next, we analyzed the temporal dynamics of these two active histone marks at attenuated (PtaZFP2, PtaXET6, and PtaACA13) and non-attenuated (PtaHRD) mechano-responsive loci during the desensitization and resensitization phases. Enrichment of H3K9/14ac and H3K4me3 in the regulatory region of attenuated genes correlates well with their transient expression pattern after the first bending. Moreover, the levels of H3K4me3 correlate well with their expression pattern after the second bending at desensitization (3 days after the first bending) as well as resensitization (5 days after the first bending) phases. On the other hand, H3K9/14ac status correlates only with their attenuated expression pattern at the desensitization phase. The expression efficiency of the attenuated genes was restored after the second bending in the histone deacetylase inhibitor-treated plants. While both histone modifications contribute to the expression of attenuated genes, mechanostimulated expression of the non-attenuated PtaHRD gene seems to be H3K4me3 dependent.</p>',
'date' => '2023-02-01',
'pmid' => 'https://doi.org/10.1101%2F2023.02.12.526104',
'doi' => '10.1101/2023.02.12.526104',
'modified' => '2023-04-14 09:20:38',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4474',
'name' => 'DNA sequence and chromatin modifiers cooperate to confer epigeneticbistability at imprinting control regions.',
'authors' => 'Butz S. et al.',
'description' => '<p>Genomic imprinting is regulated by parental-specific DNA methylation of imprinting control regions (ICRs). Despite an identical DNA sequence, ICRs can exist in two distinct epigenetic states that are memorized throughout unlimited cell divisions and reset during germline formation. Here, we systematically study the genetic and epigenetic determinants of this epigenetic bistability. By iterative integration of ICRs and related DNA sequences to an ectopic location in the mouse genome, we first identify the DNA sequence features required for maintenance of epigenetic states in embryonic stem cells. The autonomous regulatory properties of ICRs further enabled us to create DNA-methylation-sensitive reporters and to screen for key components involved in regulating their epigenetic memory. Besides DNMT1, UHRF1 and ZFP57, we identify factors that prevent switching from methylated to unmethylated states and show that two of these candidates, ATF7IP and ZMYM2, are important for the stability of DNA and H3K9 methylation at ICRs in embryonic stem cells.</p>',
'date' => '2022-11-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36333500',
'doi' => '10.1038/s41588-022-01210-z',
'modified' => '2022-11-18 12:20:16',
'created' => '2022-11-15 09:26:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4345',
'name' => 'Altered Chromatin States Drive Cryptic Transcription in AgingMammalian Stem Cells.',
'authors' => 'McCauley Brenna S et al.',
'description' => '<p>A repressive chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation suppresses cryptic transcription in embryonic stem cells. Cryptic transcription is elevated with age in yeast and nematodes, and reducing it extends yeast lifespan, though whether this occurs in mammals is unknown. We show that cryptic transcription is elevated in aged mammalian stem cells, including murine hematopoietic stem cells (mHSCs) and neural stem cells (NSCs) and human mesenchymal stem cells (hMSCs). Precise mapping allowed quantification of age-associated cryptic transcription in hMSCs aged . Regions with significant age-associated cryptic transcription have a unique chromatin signature: decreased H3K36me3 and increased H3K4me1, H3K4me3, and H3K27ac with age. Genomic regions undergoing such changes resemble known promoter sequences and are bound by TBP even in young cells. Hence, the more permissive chromatin state at intragenic cryptic promoters likely underlies increased cryptic transcription in aged mammalian stem cells.</p>',
'date' => '2021-08-01',
'pmid' => 'https://doi.org/10.1038%2Fs43587-021-00091-x',
'doi' => '10.1038/s43587-021-00091-x',
'modified' => '2022-06-22 12:30:19',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '4384',
'name' => 'Age-associated cryptic transcription in mammalian stem cells is linked topermissive chromatin at cryptic promoters',
'authors' => 'McCauley B. S. et al.',
'description' => '<p>Suppressing spurious cryptic transcription by a repressive intragenic chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation is critical for maintaining self-renewal capacity in mouse embryonic stem cells. In yeast and nematodes, such cryptic transcription is elevated with age, and reducing the levels of age-associated cryptic transcription extends yeast lifespan. Whether cryptic transcription is also increased during mammalian aging is unknown. We show for the first time an age-associated elevation in cryptic transcription in several stem cell populations, including murine hematopoietic stem cells (mHSCs) and neural stem cells (NSCs) and human mesenchymal stem cells (hMSCs). Using DECAP-seq, we mapped and quantified age-associated cryptic transcription in hMSCs aged in vitro. Regions with significant age-associated cryptic transcription have a unique chromatin signature: decreased H3K36me3 and increased H3K4me1, H3K4me3, and H3K27ac with age. Furthermore, genomic regions undergoing such age-dependent chromatin changes resemble known promoter sequences and are bound by the promoter-associated protein TBP even in young cells. Hence, the more permissive chromatin state at intragenic cryptic promoters likely underlies the increase of cryptic transcription in aged mammalian stem cells.</p>',
'date' => '2020-10-01',
'pmid' => 'https://europepmc.org/article/ppr/ppr221829',
'doi' => '10.21203/rs.3.rs-82156/v1',
'modified' => '2022-08-04 16:24:46',
'created' => '2022-08-04 14:55:36',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '3990',
'name' => 'Identification of Novel Molecular Markers of Human Th17 Cells.',
'authors' => 'Sałkowska A, Karaś K, Karwaciak I, Walczak-Drzewiecka A, Krawczyk M, Sobalska-Kwapis M, Dastych J, Ratajewski M',
'description' => '<p>Th17 cells are important players in host defense against pathogens such as , , and . Th17 cell-mediated inflammation, under certain conditions in which balance in the immune system is disrupted, is the underlying pathogenic mechanism of certain autoimmune disorders, e.g., rheumatoid arthritis, Graves' disease, multiple sclerosis, and psoriasis. In the present study, using transcriptomic profiling, we selected genes and analyzed the expression of these genes to find potential novel markers of Th17 lymphocytes. We found that (apolipoprotein D); (complement component 1, Q subcomponent-like protein 1); and (cathepsin L) are expressed at significantly higher mRNA and protein levels in Th17 cells than in the Th1, Th2, and Treg subtypes. Interestingly, these genes and the proteins they encode are well associated with the function of Th17 cells, as these cells produce inflammation, which is linked with atherosclerosis and angiogenesis. Furthermore, we found that high expression of these genes in Th17 cells is associated with the acetylation of H2BK12 within their promoters. Thus, our results provide new information regarding this cell type. Based on these results, we also hope to better identify pathological conditions of clinical significance caused by Th17 cells.</p>',
'date' => '2020-07-03',
'pmid' => 'http://www.pubmed.gov/32635226',
'doi' => '10.3390/cells9071611',
'modified' => '2020-09-01 15:05:28',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '3695',
'name' => 'The Chromatin Factor HNI9 and ELONGATED HYPOCOTYL5 Maintain ROS Homeostasis under High Nitrogen Provision.',
'authors' => 'Bellegarde F, Maghiaoui A, Boucherez J, Krouk G, Lejay L, Bach L, Gojon A, Martin A',
'description' => '<p>Reactive oxygen species (ROS) can accumulate in cells at excessive levels, leading to unbalanced redox states and to potential oxidative stress, which can have damaging effects on the molecular components of plant cells. Several environmental conditions have been described as causing an elevation of ROS production in plants. Consequently, activation of detoxification responses is necessary to maintain ROS homeostasis at physiological levels. Misregulation of detoxification systems during oxidative stress can ultimately cause growth retardation and developmental defects. Here, we demonstrate that Arabidopsis () plants grown in a high nitrogen (N) environment express a set of genes involved in detoxification of ROS that maintain ROS at physiological levels. We show that the chromatin factor HIGH NITROGEN INSENSITIVE9 (HNI9) is an important mediator of this response and is required for the expression of detoxification genes. Mutation in HNI9 leads to elevated ROS levels and ROS-dependent phenotypic defects under high but not low N provision. In addition, we identify ELONGATED HYPOCOTYL5 as a major transcription factor required for activation of the detoxification program under high N. Our results demonstrate the requirement of a balance between N metabolism and ROS production, and our work establishes major regulators required to control ROS homeostasis under conditions of excess N.</p>',
'date' => '2019-05-01',
'pmid' => 'http://www.pubmed.gov/30824566',
'doi' => '10.1104/pp.18.01473',
'modified' => '2019-06-28 13:46:23',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '3579',
'name' => 'Polycomb Repressive Complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1.',
'authors' => 'Bellegarde F, Herbert L, Séré D, Caillieux E, Boucherez J, Fizames C, Roudier F, Gojon A, Martin A',
'description' => '<p>PRC2 is a major regulator of gene expression in eukaryotes. It catalyzes the repressive chromatin mark H3K27me3, which leads to very low expression of target genes. NRT2.1, which encodes a key root nitrate transporter in Arabidopsis, is targeted by H3K27me3, but the function of PRC2 on NRT2.1 remains unclear. Here, we demonstrate that PRC2 directly targets and down-regulates NRT2.1, but in a context of very high transcription, in nutritional conditions where this gene is one of the most highly expressed genes in the transcriptome. Indeed, the mutation of CLF, which encodes a PRC2 subunit, leads to a loss of H3K27me3 at NRT2.1 and results, exclusively under permissive conditions for NRT2.1, in a further increase in NRT2.1 expression, and specifically in tissues where NRT2.1 is normally expressed. Therefore, our data indicates that PRC2 tempers the hyperactivity of NRT2.1 in a context of very strong transcription. This reveals an original function of PRC2 in the control of the expression of a highly expressed gene in Arabidopsis.</p>',
'date' => '2018-05-21',
'pmid' => 'http://www.pubmed.gov/29784958',
'doi' => '10.1038/s41598-018-26349-w',
'modified' => '2019-04-17 15:55:09',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '3377',
'name' => 'Interplay of cell–cell contacts and RhoA/MRTF‐A signaling regulates cardiomyocyte identity',
'authors' => 'Dorn et al',
'description' => '<p><span>Cell–cell and cell–matrix interactions guide organ development and homeostasis by controlling lineage specification and maintenance, but the underlying molecular principles are largely unknown. Here, we show that in human developing cardiomyocytes cell–cell contacts at the intercalated disk connect to remodeling of the actin cytoskeleton by regulating the RhoA‐</span><span class="styled-content fixed-case">ROCK</span><span><span> </span>signaling to maintain an active<span> </span></span><span class="styled-content fixed-case">MRTF</span><span>/</span><span class="styled-content fixed-case">SRF</span><span><span> </span>transcriptional program essential for cardiomyocyte identity. Genetic perturbation of this mechanosensory pathway activates an ectopic fat gene program during cardiomyocyte differentiation, which ultimately primes the cells to switch to the brown/beige adipocyte lineage in response to adipogenesis‐inducing signals. We also demonstrate by<span> </span></span><em>in vivo</em><span><span> </span>fate mapping and clonal analysis of cardiac progenitors that cardiac fat and a subset of cardiac muscle arise from a common precursor expressing Isl1 and Wt1 during heart development, suggesting related mechanisms of determination between the two lineages.</span></p>',
'date' => '2018-05-15',
'pmid' => 'http://emboj.embopress.org/content/early/2018/05/15/embj.201798133',
'doi' => '10.15252/embj.201798133',
'modified' => '2018-05-22 22:50:53',
'created' => '2018-05-22 22:50:53',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '3473',
'name' => 'Neonatal exposure to hyperoxia leads to persistent disturbances in pulmonary histone signatures associated with NOS3 and STAT3 in a mouse model.',
'authors' => 'Chao CM, van den Bruck R, Lork S, Merkle J, Krampen L, Weil PP, Aydin M, Bellusci S, Jenke AC, Postberg J',
'description' => '<p>Background: Early pulmonary oxygen exposure is one of the most important factors implicated in the development of bronchopulmonary dysplasia (BPD). Methods: Here, we analyzed short- and long-term effects of neonatal hyperoxia on NOS3 and STAT3 expression and corresponding epigenetic signatures using a hyperoxia-based mouse model of BPD. Results: Early hyperoxia exposure led to a significant increase in NOS3 (median fold change × 2.37, IQR 1.54-3.68) and STAT3 (median fold change × 2.83, IQR 2.21-3.88) mRNA levels in pulmonary endothelial cells with corresponding changes in histone modification patterns such as H2aZac and H3K9ac hyperacetylation at the respective gene loci. No complete restoration in histone signatures at these loci was observed, and responsivity to later hyperoxia was altered in mouse lungs. In vitro, histone signatures in human aortic endothelial cells (HAEC) remained altered for several weeks after an initial long-term exposure to trichostatin A. This was associated with a substantial increase in baseline eNOS (median 27.2, IQR 22.3-35.6) and STAT3α (median 5.8, IQR 4.8-7.3) mRNA levels with a subsequent significant reduction in eNOS expression upon exposure to hypoxia. Conclusions: Early hyperoxia induced permanent changes in histones signatures at the NOS3 and STAT3 gene locus might partly explain the altered vascular response patterns in children with BPD.</p>',
'date' => '2018-01-01',
'pmid' => 'http://www.pubmed.gov/29581793',
'doi' => '10.1186/s13148-018-0469-0',
'modified' => '2019-02-15 21:45:12',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '3299',
'name' => 'Rapid Communication: The correlation between histone modifications and expression of key genes involved in accumulation of adipose tissue in the pig.',
'authors' => 'Kociucka B. et al.',
'description' => '<p>Histone modification is a well-known epigenetic mechanism involved in regulation of gene expression; however, it has been poorly studied in adipose tissues of the pig. Understanding the molecular background of adipose tissue development and function is essential for improving production efficiency and meat quality. The objective of this study was to identify the association between histone modification and the transcript level of genes important for lipid droplet formation and metabolism. Histone modifications at the promoter regions of 6 genes (, , , , , and ) were analyzed using a chromatin immunoprecipitation assay. Two modifications involved in activation of gene expression (acetylation of H3 histone at lysine 9 and methylation of H3 histone at lysine 4) as well as methylation of H3 histone at lysine 27, which is known to be related to gene repression, were examined. The level of histone modification was compared with transcript abundance determined using real-time PCR in tissue samples (subcutaneous fat, visceral fat, and longissimus dorsi muscle) derived from 3 pig breeds significantly differing in fatness traits (Polish Large White, Duroc, and Pietrain). Transcript levels were found to be correlated with histone modifications characteristic to active loci in 4 of 6 genes. A positive correlation between histone H3 lysine 9 acetylation modification and the transcript level of ( = 0.53, < 4.8 × 10), ( = 0.34, < 0.02), and ( = 0.43, < 1.0 × 10) genes was observed. The histone H3 lysine 4 trimethylation modification correlated with transcripts of ( = 0.64, < 4.6 × 10) and ( = 0.37, < 0.01) genes. No correlation was found between transcript level of all studied genes and histone H3 lysine 27 trimethylation level. This is the first study on histone modifications in porcine adipose tissues. We confirmed the relationship between histone modifications and expression of key genes for adipose tissue accumulation in the pig. Epigenetic modulation of the transcriptional profile of these genes (e.g., through nutritional factors) may improve porcine fatness traits in future.</p>',
'date' => '2017-10-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29108067',
'doi' => '',
'modified' => '2017-12-05 10:39:56',
'created' => '2017-12-05 09:31:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '3083',
'name' => 'Lhx2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11',
'authors' => 'Muralidharan B. et al.',
'description' => '<p>n the developing cerebral cortex, sequential transcriptional programs take neuroepithelial cells from proliferating progenitors to differentiated neurons with unique molecular identities. The regulatory changes that occur in the chromatin of the progenitors are not well understood. During deep layer neurogenesis, we show that transcription factor Lhx2 binds to distal regulatory elements of Fezf2 and Sox11, critical determinants of neuron subtype identity in the mouse neocortex. We demonstrate that Lhx2 binds to the NuRD histone remodeling complex subunits LSD1, HDAC2, and RBBP4, which are proximal regulators of the epigenetic state of chromatin. When Lhx2 is absent, active histone marks at the Fezf2 and Sox11 loci are increased. Loss of Lhx2 produces an increase, and overexpression of Lhx2 causes a decrease, in layer 5 Fezf2 and Ctip2 expressing neurons. Our results provide mechanistic insight into how Lhx2 acts as a necessary and sufficient regulator of genes that control cortical neuronal subtype identity.</p>',
'date' => '2016-12-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27909100',
'doi' => '',
'modified' => '2016-12-20 10:28:02',
'created' => '2016-12-20 10:28:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '2910',
'name' => 'Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape',
'authors' => 'Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">Previously, a role was demonstrated for transcription in the acquisition of DNA methylation at imprinted control regions in oocytes. Definition of the oocyte DNA methylome by whole genome approaches revealed that the majority of methylated CpG islands are intragenic and gene bodies are hypermethylated. Yet, the mechanisms by which transcription regulates DNA methylation in oocytes remain unclear. Here, we systematically test the link between transcription and the methylome.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We perform deep RNA-Seq and de novo transcriptome assembly at different stages of mouse oogenesis. This reveals thousands of novel non-annotated genes, as well as alternative promoters, for approximately 10 % of reference genes expressed in oocytes. In addition, a large fraction of novel promoters coincide with MaLR and ERVK transposable elements. Integration with our transcriptome assembly reveals that transcription correlates accurately with DNA methylation and accounts for approximately 85-90 % of the methylome. We generate a mouse model in which transcription across the Zac1/Plagl1 locus is abrogated in oocytes, resulting in failure of DNA methylation establishment at all CpGs of this locus. ChIP analysis in oocytes reveals H3K4me2 enrichment at the Zac1 imprinted control region when transcription is ablated, establishing a connection between transcription and chromatin remodeling at CpG islands by histone demethylases.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">By precisely defining the mouse oocyte transcriptome, this work not only highlights transcription as a cornerstone of DNA methylation establishment in female germ cells, but also provides an important resource for developmental biology research.</abstracttext></p>
</div>',
'date' => '2015-09-25',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26408185',
'doi' => '10.1186/s13059-015-0769-z',
'modified' => '2016-05-10 22:01:49',
'created' => '2016-05-10 22:01:49',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '2613',
'name' => 'CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.',
'authors' => 'Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC',
'description' => 'BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the underlying epigenetic signatures involved in its regulation. RESULTS: NOS3 and STAT3 mRNA levels were elevated in HUAEC of patients who suffered from placental insufficiency. 5-hydroxymethylcytosine, H3K9ac and Histone 2A (H2A).Zac at the NOS3 transcription start site directly correlated with NOS3 mRNA levels. Concomitantly, we observed entangled histone acetylation patterns and NOS3 response upon hypoxic conditions in vitro. Knock-down of either NOS3 or STAT3 by RNAi provided evidence for a functional NOS3/STAT3 relationship. Moreover, we recognized massive turnover of Stat3 at a discrete binding site in the NOS3 promoter. Interestingly, induced hyperacetylation resulted in short-termed increase of NOS3 mRNA followed by deferred decrease indicating that NOS3 expression could become self-attenuated by co-expressed intronic 27 nt-ncRNA. Reporter assay results and phylogenetic analyses enabled us to propose a novel model for STAT3-3'-UTR targeting by this 27-nt-ncRNA. CONCLUSIONS: An adverse intrauterine environment leads to adaptive changes of NOS3 expression. Apparently, a rapid NOS3 self-limiting response upon ectopic triggers co-exists with longer termed expression changes in response to placental insufficiency involving differential epigenetic signatures. Their persistence might contribute to impaired vascular endothelial response and consequently increase the risk of cardiovascular disease later in life.',
'date' => '2015-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25699114',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '2506',
'name' => 'Paclitaxel resistance increases oncolytic adenovirus efficacy via upregulated CAR expression and dysfunctional cell cycle control.',
'authors' => 'Ingemarsdotter CK, Tookman LA, Browne A, Pirlo K, Cutts R, Chelela C, Khurrum KF, Leung EY, Dowson S, Webber L, Khan I, Ennis D, Syed N, Crook TR, Brenton JD, Lockley M, McNeish IA',
'description' => 'Resistance to paclitaxel chemotherapy frequently develops in ovarian cancer. Oncolytic adenoviruses are a novel therapy for human malignancies that are being evaluated in early phase trials. However, there are no reliable predictive biomarkers for oncolytic adenovirus activity in ovarian cancer. We investigated the link between paclitaxel resistance and oncolytic adenovirus activity using established ovarian cancer cell line models, xenografts with de novo paclitaxel resistance and tumour samples from two separate trials. The activity of multiple Ad5 vectors, including dl922-947 (E1A CR2-deleted), dl1520 (E1B-55K deleted) and Ad5 WT, was significantly increased in paclitaxel resistant ovarian cancer in vitro and in vivo. This was associated with greater infectivity resulting from increased expression of the primary receptor for Ad5, CAR (coxsackie adenovirus receptor). This, in turn, resulted from increased CAR transcription secondary to histone modification in resistant cells. There was increased CAR expression in intraperitoneal tumours with de novo paclitaxel resistance and in tumours from patients with clinical resistance to paclitaxel. Increased CAR expression did not cause paclitaxel resistance, but did increase inflammatory cytokine expression. Finally, we identified dysregulated cell cycle control as a second mechanism of increased adenovirus efficacy in paclitaxel-resistant ovarian cancer. Ad11 and Ad35, both group B adenoviruses that utilise non-CAR receptors to infect cells, are also significantly more effective in paclitaxel-resistant ovarian cell models. Inhibition of CDK4/6 using PD-0332991 was able both to reverse paclitaxel resistance and reduce adenovirus efficacy. Thus, paclitaxel resistance increases oncolytic adenovirus efficacy via at least two separate mechanisms - if validated further, this information could have future clinical utility to aid patient selection for clinical trials.',
'date' => '2014-12-29',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25560085',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '998',
'name' => 'p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6.',
'authors' => 'Phalke S, Mzoughi S, Bezzi M, Jennifer N, Mok WC, Low DH, Thike AA, Kuznetsov VA, Tan PH, Voorhoeve PM, Guccione E',
'description' => 'p21 is a potent cyclin-dependent kinase inhibitor that plays a role in promoting G1 cell cycle arrest and cellular senescence. Consistent with this role, p21 is a downstream target of several tumour suppressors and oncogenes, and it is downregulated in the majority of tumours, including breast cancer. Here, we report that protein arginine methyltransferase 6 (PRMT6), a type I PRMT known to act as a transcriptional cofactor, directly represses the p21 promoter. PRMT6 knock-down (KD) results in a p21 derepression in breast cancer cells, which is p53-independent, and leads to cell cycle arrest, cellular senescence and reduced growth in soft agar assays and in severe combined immunodeficiency (SCID) mice for all the cancer lines examined. We finally show that bypassing the p21-mediated arrest rescues PRMT6 KD cells from senescence, and it restores their ability to grow on soft agar. We conclude that PRMT6 acts as an oncogene in breast cancer cells, promoting growth and preventing senescence, making it an attractive target for cancer therapy.',
'date' => '2012-09-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22987071',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '359',
'name' => 'Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.',
'authors' => 'Henckel A, Chebli K, Kota SK, Arnaud P, Feil R',
'description' => 'Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at 'maternal' ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.',
'date' => '2012-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22117218',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '432',
'name' => 'H3K4me3 antibody SDS US en',
'language' => 'en',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-US-en-GHS_2_0.pdf',
'countries' => 'US',
'modified' => '2020-06-30 18:23:32',
'created' => '2020-06-30 18:23:32',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '430',
'name' => 'H3K4me3 antibody SDS GB en',
'language' => 'en',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-GB-en-GHS_2_0.pdf',
'countries' => 'GB',
'modified' => '2020-06-30 18:22:36',
'created' => '2020-06-30 18:22:36',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '425',
'name' => 'H3K4me3 antibody SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-fr-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:17:31',
'created' => '2020-06-30 18:17:31',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '429',
'name' => 'H3K4me3 antibody SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-30 18:22:02',
'created' => '2020-06-30 18:22:02',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '428',
'name' => 'H3K4me3 antibody SDS ES es',
'language' => 'es',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-ES-es-GHS_2_0.pdf',
'countries' => 'ES',
'modified' => '2020-06-30 18:21:17',
'created' => '2020-06-30 18:21:17',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '427',
'name' => 'H3K4me3 antibody SDS DE de',
'language' => 'de',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-DE-de-GHS_2_0.pdf',
'countries' => 'DE',
'modified' => '2020-06-30 18:19:09',
'created' => '2020-06-30 18:19:09',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '431',
'name' => 'H3K4me3 antibody SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-JP-ja-GHS_2_0.pdf',
'countries' => 'JP',
'modified' => '2020-06-30 18:23:02',
'created' => '2020-06-30 18:23:02',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '426',
'name' => 'H3K4me3 antibody SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:18:22',
'created' => '2020-06-30 18:18:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/h3k4me3-polyclonal-antibody-classic-50-ug-25-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
)
$pro = array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/bioruptor-pico-sonication-device"><img src="/img/product/shearing_technologies/bioruptor_pico.jpg" alt="Bioruptor pico next gen sequencing " class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">B01060010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Bioruptor® Pico sonication device</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '1101',
'product_id' => '2195',
'related_id' => '1787'
),
'Image' => array(
(int) 0 => array(
'id' => '133',
'name' => 'product/shearing_technologies/bioruptor_pico.jpg',
'alt' => 'Bioruptor pico next gen sequencing ',
'modified' => '2020-11-27 10:00:30',
'created' => '2015-06-10 17:28:55',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1772',
'name' => 'product/shearing_technologies/B010600010.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 15:41:46',
'created' => '2018-02-14 15:41:46',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1773',
'name' => 'product/shearing_technologies/B010600010-2.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 16:18:03',
'created' => '2018-02-14 15:42:00',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1774',
'name' => 'product/shearing_technologies/B01060010-bioruptor-pico-cooler.jpg',
'alt' => 'Pico + cooler',
'modified' => '2018-03-06 11:01:33',
'created' => '2018-03-06 11:01:33',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'ProductsApplication' => array(
'id' => '4314',
'product_id' => '2195',
'application_id' => '42'
)
)
$slugs = array(
(int) 0 => 'chip-seq-antibodies'
)
$applications = array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'locale' => 'eng'
)
$description = ''
$name = 'ChIP-seq (ab)'
$document = array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '1616',
'product_id' => '2195',
'document_id' => '38'
)
)
$sds = array(
'id' => '426',
'name' => 'H3K4me3 antibody SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:18:22',
'created' => '2020-06-30 18:18:22',
'ProductsSafetySheet' => array(
'id' => '833',
'product_id' => '2195',
'safety_sheet_id' => '426'
)
)
$publication = array(
'id' => '359',
'name' => 'Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.',
'authors' => 'Henckel A, Chebli K, Kota SK, Arnaud P, Feil R',
'description' => 'Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at 'maternal' ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.',
'date' => '2012-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22117218',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
'id' => '870',
'product_id' => '2195',
'publication_id' => '359'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/22117218" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
Notice (8): Undefined variable: campaign_id [APP/View/Products/view.ctp, line 755]Code Context<!-- BEGIN: REQUEST_FORM MODAL -->
<div id="request_formModal" class="reveal-modal medium" data-reveal aria-labelledby="modalTitle" aria-hidden="true" role="dialog">
<?= $this->element('Forms/simple_form', array('solution_of_interest' => $solution_of_interest, 'header' => $header, 'message' => $message, 'campaign_id' => $campaign_id)) ?>
$viewFile = '/home/website-server/www/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'product' => array(
'Product' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '114',
'name' => 'H3K4me3 polyclonal antibody',
'description' => 'Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.',
'clonality' => '',
'isotype' => '',
'lot' => '002',
'concentration' => '1.4 µg/µl',
'reactivity' => 'Human, mouse, Arabidopsis: positive. Other species: not tested.',
'type' => 'Polyclonal',
'purity' => 'Affinity purified polyclonal antibody.',
'classification' => 'Classic',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>ChIP/ChIP-seq <sup>*</sup></td>
<td>1 µg/ChIP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting</td>
<td>1:2,000</td>
<td>Fig 3</td>
</tr>
<tr>
<td>Western Blotting</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
<tr>
<td>Immunofluorescence</td>
<td>1:100</td>
<td>Fig 5</td>
</tr>
</tbody>
</table>
<p></p>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 µg per IP.</small></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide and 0.05% ProClin 300.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-11-25 17:04:17',
'created' => '0000-00-00 00:00:00',
'select_label' => '114 - H3K4me3 polyclonal antibody (002 - 1.4 µg/µl - Human, mouse, Arabidopsis: positive. Other species: not tested. - Affinity purified polyclonal antibody. - Rabbit)'
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Application' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
)
),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://www.diagenode.com/en/p/h3k4me3-polyclonal-antibody-classic-50-ug-25-ul'
)
$language = 'en'
$meta_keywords = 'H3K4me3 Antibody '
$meta_description = 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.'
$meta_title = 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode'
$product = array(
'Product' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => '114',
'name' => 'H3K4me3 polyclonal antibody',
'description' => 'Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.',
'clonality' => '',
'isotype' => '',
'lot' => '002',
'concentration' => '1.4 µg/µl',
'reactivity' => 'Human, mouse, Arabidopsis: positive. Other species: not tested.',
'type' => 'Polyclonal',
'purity' => 'Affinity purified polyclonal antibody.',
'classification' => 'Classic',
'application_table' => '<table>
<thead>
<tr>
<th>Applications</th>
<th>Suggested dilution</th>
<th>References</th>
</tr>
</thead>
<tbody>
<tr>
<td>ChIP/ChIP-seq <sup>*</sup></td>
<td>1 µg/ChIP</td>
<td>Fig 1, 2</td>
</tr>
<tr>
<td>Dot Blotting</td>
<td>1:2,000</td>
<td>Fig 3</td>
</tr>
<tr>
<td>Western Blotting</td>
<td>1:500</td>
<td>Fig 4</td>
</tr>
<tr>
<td>Immunofluorescence</td>
<td>1:100</td>
<td>Fig 5</td>
</tr>
</tbody>
</table>
<p></p>
<p><small><sup>*</sup> Please note that the optimal antibody amount per IP should be determined by the end-user. We recommend testing 1-5 µg per IP.</small></p>',
'storage_conditions' => 'Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles.',
'storage_buffer' => 'PBS containing 0.05% azide and 0.05% ProClin 300.',
'precautions' => 'This product is for research use only. Not for use in diagnostic or therapeutic procedures.',
'uniprot_acc' => '',
'slug' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2020-11-25 17:04:17',
'created' => '0000-00-00 00:00:00',
'select_label' => '114 - H3K4me3 polyclonal antibody (002 - 1.4 µg/µl - Human, mouse, Arabidopsis: positive. Other species: not tested. - Affinity purified polyclonal antibody. - Rabbit)'
),
'Slave' => array(
(int) 0 => array(
'id' => '322',
'name' => 'C15410030',
'product_id' => '2195',
'modified' => '2020-11-25 16:06:48',
'created' => '2020-11-25 16:06:48'
)
),
'Group' => array(
'Group' => array(
'id' => '322',
'name' => 'C15410030',
'product_id' => '2195',
'modified' => '2020-11-25 16:06:48',
'created' => '2020-11-25 16:06:48'
),
'Master' => array(
'id' => '2195',
'antibody_id' => '114',
'name' => 'H3K4me3 Antibody',
'description' => '<p>Polyclonal antibody raised in rabbit against the region of <strong>histone H3</strong> containing the <strong>trimethylated lysine 4</strong> (<strong>H3K4me3</strong>), using a KLH-conjugated synthetic peptide.</p>',
'label1' => 'Validation data',
'info1' => '<div class="row">
<div class="small-6 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-1.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chip-2.jpg" alt="H3K4me3 Antibody ChIP Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 1. ChIP results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP assays were performed using human HeLa cells, the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and optimized PCR primer pairs for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (cat. No. C01010051), using sheared chromatin from 1 million cells. The chromatin was spiked with a panel of in vitro assembled nucleosomes, each containing a specific lysine methylation (SNAP-ChIP K-MetStat Panel, Epicypher). A titration consisting of 1, 2, 5 and 10 µg of antibody per ChIP experiment was analyzed. IgG (2 µg/IP) was used as a negative IP control. <strong>Figure 1A.</strong> Quantitative PCR was performed with primers specific for the promoter of the active GAPDH and EIF4A2 genes, used as positive controls, and for the inactive MYOD1 gene, used as negative control. The graph shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis). These results are in accordance with the observation that trimethylation of K4 at histone H3 is associated with the promoters of active genes <strong>Figure 1B.</strong> Recovery of the nucleosomes carrying the H3K4me1, H3K4me2, H3K4me3 modifications and the unmodified H3K4 as determined by qPCR. The figure clearly shows the antibody is very specific in ChIP for the H3K4me3 modification. At higher concentrations some H3K4me2 is also precipitated. </small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p>A. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-a.jpg" alt="H3K4me3 Antibody ChIP-seq Grade" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>B. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-b.jpg" alt="H3K4me3 Antibody for ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<p>C. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-c.jpg" alt="H3K4me3 Antibody for ChIP-seq assay" style="display: block; margin-left: auto; margin-right: auto;" /></p>
<p>D. <img src="https://www.diagenode.com/img/product/antibodies/c15410030-chipseq-d.jpg" alt="H3K4me3 Antibody validated in ChIP-seq" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against H3K4me3 </strong><br />ChIP was performed on sheared chromatin from 1 million HeLa cells using 1 µg of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) as described above. The IP'd DNA was subsequently analysed on an Illumina Genome Analyzer. Library preparation, cluster generation and sequencing were performed according to the manufacturer's instructions. The 50 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the complete sequence and a 1.2 Mb region of the X-chromosome (figure 2A and B) and in two regions surrounding the GAPDH and EIF4A2 positive control genes, respectively (figure 2C and D). These results clearly show an enrichment of the H3K4 trimethylation at the promoters of active genes.</small></p>
</div>
</div>
<div class="row">
<div class="small-6 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-dotblot.jpg" alt="H3K4me3 Antibody validated in Dot Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-6 columns">
<p><small><strong> Figure 3. Cross reactivity test using the Diagenode antibody directed against H3K4me3 </strong><br />A Dot Blot analysis was performed to test the cross reactivity of the Diagenode antibody against H3K4me3 (Cat. No. C15410030) with peptides containing other modifications and unmodified sequences of histone H3 and H4. One hundred to 0.2 pmol of the respective peptides were spotted on a membrane. The antibody was used at a dilution of 1:2,000. Figure 3 shows a high specificity of the antibody for the modification of interest.</small></p>
</div>
</div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="extra-spaced"></div>
<div class="row">
<div class="small-4 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-wb.jpg" alt="H3K4me3 Antibody validated in Western Blot" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
<div class="small-8 columns">
<p><small><strong> Figure 4. Western blot analysis using the Diagenode antibody directed against H3K4me3 </strong><br />Western blot was performed on whole cell (40 µg, lane 1) and histone extracts (15 µg, lane 2) from HeLa cells, and on 1 µg of recombinant histone H2A, H2B, H3.1, H3.3 and H4 (lane 3, 4, 5, 6 and 7, respectively) using the Diagenode antibody against H3K4me3 (Cat. No. C15410030). The antibody was diluted 1:500 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest is shown on the right, the marker (in kDa) is shown on the left.</small></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><img src="https://www.diagenode.com/img/product/antibodies/c15410030-if.jpg" alt="H3K4me3 Antibody validated in Immunofluorescence" style="display: block; margin-left: auto; margin-right: auto;" /></p>
</div>
</div>
<div class="row">
<div class="small-12 columns">
<p><small><strong> Figure 5. Immunofluorescence using the Diagenode antibody directed against H3K4me3 </strong><br />HeLa cells were stained with the Diagenode antibody against H3K4me3 (Cat. No. C15410030) and with DAPI. Cells were fixed with 4% formaldehyde for 10’ and blocked with PBS/TX-100 containing 5% normal goat serum and 1% BSA. The cells were immunofluorescently labelled with the H3K4me3 antibody (left) diluted 1:100 in blocking solution followed by an anti-rabbit antibody conjugated to Alexa488. The middle panel shows staining of the nuclei with DAPI. A merge of the two stainings is shown on the right.</small></p>
</div>
</div>',
'label2' => 'Target Description',
'info2' => '<p>Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. In addition to the genetic code, combinations of the different histone modifications reveal the so-called “histone code”. Histone methylation and demethylation is dynamically regulated by respectively histone methyl transferases and histone demethylases. Trimethylation of histone H3K4 is associated with active promoters.</p>',
'label3' => '',
'info3' => '',
'format' => '50 µg',
'catalog_number' => 'C15410030',
'old_catalog_number' => 'pAb-030-050',
'sf_code' => 'C15410030-D001-000581',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '380',
'price_USD' => '380',
'price_GBP' => '340',
'price_JPY' => '59525',
'price_CNY' => '',
'price_AUD' => '950',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-50-ug-25-ul',
'meta_title' => 'H3K4me3 Antibody - ChIP Grade (C15410030) | Diagenode',
'meta_keywords' => 'H3K4me3 Antibody ',
'meta_description' => 'H3K4me3 Antibody validated in ChIP-seq, ChIP-qPCR, DB, WB and IF. Batch-specific data available on the website.',
'modified' => '2021-12-23 11:31:39',
'created' => '2015-06-29 14:08:20'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(
(int) 0 => array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
[maximum depth reached]
),
'Image' => array(
[maximum depth reached]
)
)
),
'Application' => array(
(int) 0 => array(
'id' => '28',
'position' => '10',
'parent_id' => '40',
'name' => 'DB',
'description' => '<p>Dot blotting</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'dot-blotting',
'meta_keywords' => 'Dot blotting,Monoclonal & Polyclonal antibody,',
'meta_description' => 'Diagenode offers Monoclonal & Polyclonal antibodies for Dot blotting applications',
'meta_title' => 'Dot blotting Antibodies - Monoclonal & Polyclonal antibody | Diagenode',
'modified' => '2016-01-13 14:40:49',
'created' => '2015-07-08 13:45:05',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '19',
'position' => '10',
'parent_id' => '40',
'name' => 'WB',
'description' => '<p><strong>Western blot</strong> : The quality of antibodies used in this technique is crucial for correct and specific protein identification. Diagenode offers huge selection of highly sensitive and specific western blot-validated antibodies.</p>
<p>Learn more about: <a href="https://www.diagenode.com/applications/western-blot">Loading control, MW marker visualization</a><em>. <br /></em></p>
<p><em></em>Check our selection of antibodies validated in Western blot.</p>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'western-blot-antibodies',
'meta_keywords' => ' Western Blot Antibodies ,western blot protocol,Western Blotting Products,Polyclonal antibodies ,monoclonal antibodies ',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for western blot applications',
'meta_title' => ' Western Blot - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-26 12:44:51',
'created' => '2015-01-07 09:20:00',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '29',
'position' => '10',
'parent_id' => '40',
'name' => 'IF',
'description' => '<p><strong>Immunofluorescence</strong>:</p>
<p>Diagenode offers huge selection of highly sensitive antibodies validated in IF.</p>
<p><img src="https://www.diagenode.com/img/product/antibodies/C15200229-IF.jpg" alt="" height="245" width="256" /></p>
<p><sup><strong>Immunofluorescence using the Diagenode monoclonal antibody directed against CRISPR/Cas9</strong></sup></p>
<p><sup>HeLa cells transfected with a Cas9 expression vector (left) or untransfected cells (right) were fixed in methanol at -20°C, permeabilized with acetone at -20°C and blocked with PBS containing 2% BSA. The cells were stained with the Cas9 C-terminal antibody (Cat. No. C15200229) diluted 1:400, followed by incubation with an anti-mouse secondary antibody coupled to AF488. The bottom images show counter-staining of the nuclei with Hoechst 33342.</sup></p>
<h5><sup>Check our selection of antibodies validated in IF.</sup></h5>',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'immunofluorescence',
'meta_keywords' => 'Immunofluorescence,Monoclonal antibody,Polyclonal antibody',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for Immunofluorescence applications',
'meta_title' => 'Immunofluorescence - Monoclonal antibody - Polyclonal antibody | Diagenode',
'modified' => '2016-04-27 16:23:10',
'created' => '2015-07-08 13:46:02',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '43',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-qPCR (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-qpcr-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP-qPCR applications',
'meta_title' => 'ChIP Quantitative PCR Antibodies (ChIP-qPCR) | Diagenode',
'modified' => '2016-01-20 11:30:24',
'created' => '2015-10-20 11:45:36',
'ProductsApplication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'ProductsApplication' => array(
[maximum depth reached]
)
)
),
'Category' => array(
(int) 0 => array(
'id' => '111',
'position' => '40',
'parent_id' => '4',
'name' => 'Histone antibodies',
'description' => '<p>Histones are the main protein components of chromatin involved in the compaction of DNA into nucleosomes, the basic units of chromatin. A <strong>nucleosome</strong> consists of one pair of each of the core histones (<strong>H2A</strong>, <strong>H2B</strong>, <strong>H3</strong> and <strong>H4</strong>) forming an octameric structure wrapped by 146 base pairs of DNA. The different nucleosomes are linked by the linker histone<strong> H1, </strong>allowing for further condensation of chromatin.</p>
<p>The core histones have a globular structure with large unstructured N-terminal tails protruding from the nucleosome. They can undergo to multiple post-translational modifications (PTM), mainly at the N-terminal tails. These <strong>post-translational modifications </strong>include methylation, acetylation, phosphorylation, ubiquitinylation, citrullination, sumoylation, deamination and crotonylation. The most well characterized PTMs are <strong>methylation,</strong> <strong>acetylation and phosphorylation</strong>. Histone methylation occurs mainly on lysine (K) residues, which can be mono-, di- or tri-methylated, and on arginines (R), which can be mono-methylated and symmetrically or asymmetrically di-methylated. Histone acetylation occurs on lysines and histone phosphorylation mainly on serines (S), threonines (T) and tyrosines (Y).</p>
<p>The PTMs of the different residues are involved in numerous processes such as DNA repair, DNA replication and chromosome condensation. They influence the chromatin organization and can be positively or negatively associated with gene expression. Trimethylation of H3K4, H3K36 and H3K79, and lysine acetylation generally result in an open chromatin configuration (figure below) and are therefore associated with <strong>euchromatin</strong> and gene activation. Trimethylation of H3K9, K3K27 and H4K20, on the other hand, is enriched in <strong>heterochromatin </strong>and associated with gene silencing. The combination of different histone modifications is called the "<strong>histone code</strong>”, analogous to the genetic code.</p>
<p><img src="https://www.diagenode.com/img/categories/antibodies/histone-marks-illustration.png" /></p>
<p>Diagenode is proud to offer a large range of antibodies against histones and histone modifications. Our antibodies are highly specific and have been validated in many applications, including <strong>ChIP</strong> and <strong>ChIP-seq</strong>.</p>
<p>Diagenode’s collection includes antibodies recognizing:</p>
<ul>
<li><strong>Histone H1 variants</strong></li>
<li><strong>Histone H2A, H2A variants and histone H2A</strong> <strong>modifications</strong> (serine phosphorylation, lysine acetylation, lysine ubiquitinylation)</li>
<li><strong>Histone H2B and H2B</strong> <strong>modifications </strong>(serine phosphorylation, lysine acetylation)</li>
<li><strong>Histone H3 and H3 modifications </strong>(lysine methylation (mono-, di- and tri-methylated), lysine acetylation, serine phosphorylation, threonine phosphorylation, arginine methylation (mono-methylated, symmetrically and asymmetrically di-methylated))</li>
<li><strong>Histone H4 and H4 modifications (</strong>lysine methylation (mono-, di- and tri-methylated), lysine acetylation, arginine methylation (mono-methylated and symmetrically di-methylated), serine phosphorylation )</li>
</ul>
<p><span style="font-weight: 400;"><strong>HDAC's HAT's, HMT's and other</strong> <strong>enzymes</strong> which modify histones can be found in the category <a href="../categories/chromatin-modifying-proteins-histone-transferase">Histone modifying enzymes</a><br /></span></p>
<p><span style="font-weight: 400;"> Diagenode’s highly validated antibodies:</span></p>
<ul>
<li><span style="font-weight: 400;"> Highly sensitive and specific</span></li>
<li><span style="font-weight: 400;"> Cost-effective (requires less antibody per reaction)</span></li>
<li><span style="font-weight: 400;"> Batch-specific data is available on the website</span></li>
<li><span style="font-weight: 400;"> Expert technical support</span></li>
<li><span style="font-weight: 400;"> Sample sizes available</span></li>
<li><span style="font-weight: 400;"> 100% satisfaction guarantee</span></li>
</ul>',
'no_promo' => false,
'in_menu' => false,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'histone-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Histone, antibody, histone h1, histone h2, histone h3, histone h4',
'meta_description' => 'Polyclonal and Monoclonal Antibodies against Histones and their modifications validated for many applications, including Chromatin Immunoprecipitation (ChIP) and ChIP-Sequencing (ChIP-seq)',
'meta_title' => 'Histone and Modified Histone Antibodies | Diagenode',
'modified' => '2020-09-17 13:34:56',
'created' => '2016-04-01 16:01:32',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 1 => array(
'id' => '103',
'position' => '0',
'parent_id' => '4',
'name' => 'All antibodies',
'description' => '<p><span style="font-weight: 400;">All Diagenode’s antibodies are listed below. Please, use our Quick search field to find the antibody of interest by target name, application, purity.</span></p>
<p><span style="font-weight: 400;">Diagenode’s highly validated antibodies:</span></p>
<ul>
<li>Highly sensitive and specific</li>
<li>Cost-effective (requires less antibody per reaction)</li>
<li>Batch-specific data is available on the website</li>
<li>Expert technical support</li>
<li>Sample sizes available</li>
<li>100% satisfaction guarantee</li>
</ul>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'all-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'Antibodies,Premium Antibodies,Classic,Pioneer',
'meta_description' => 'Diagenode Offers Strict quality standards with Rigorous QC and validated Antibodies. Classified based on level of validation for flexibility of Application. Comprehensive selection of histone and non-histone Antibodies',
'meta_title' => 'Diagenode's selection of Antibodies is exclusively dedicated for Epigenetic Research | Diagenode',
'modified' => '2019-07-03 10:55:44',
'created' => '2015-11-02 14:49:22',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
),
(int) 2 => array(
'id' => '127',
'position' => '10',
'parent_id' => '4',
'name' => 'ChIP-grade antibodies',
'description' => '<div class="row">
<div class="small-10 columns"><center></center>
<p><br />Chromatin immunoprecipitation (<b>ChIP</b>) is a technique to study the associations of proteins with the specific genomic regions in intact cells. One of the most important steps of this protocol is the immunoprecipitation of targeted protein using the antibody specifically recognizing it. The quality of antibodies used in ChIP is essential for the success of the experiment. Diagenode offers extensively validated ChIP-grade antibodies, confirmed for their specificity, and high level of performance in ChIP. Each batch is validated, and batch-specific data are available on the website.</p>
<p></p>
</div>
<div class="small-2 columns"><img src="https://www.diagenode.com/emailing/images/epi-success-guaranteed-icon.png" alt="Epigenetic success guaranteed" /></div>
</div>
<p><strong>ChIP results</strong> obtained with the antibody directed against H3K4me3 (Cat. No. <a href="../p/h3k4me3-polyclonal-antibody-premium-50-ug-50-ul">C15410003</a>). </p>
<div class="row">
<div class="small-12 medium-6 large-6 columns"><img src="https://www.diagenode.com/img/product/antibodies/C15410003-fig1-ChIP.jpg" alt="" width="400" height="315" /> </div>
<div class="small-12 medium-6 large-6 columns">
<p></p>
<p></p>
<p></p>
</div>
</div>
<p></p>
<p>Our aim at Diagenode is to offer the largest collection of highly specific <strong>ChIP-grade antibodies</strong>. We add new antibodies monthly. Find your ChIP-grade antibody in the list below and check more information about tested applications, extensive validation data, and product information.</p>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => false,
'hide' => true,
'all_format' => false,
'is_antibody' => true,
'slug' => 'chip-grade-antibodies',
'cookies_tag_id' => null,
'meta_keywords' => 'ChIP-grade antibodies, polyclonal antibody, monoclonal antibody, Diagenode',
'meta_description' => 'Diagenode Offers Extensively Validated ChIP-Grade Antibodies, Confirmed for their Specificity, and high level of Performance in Chromatin Immunoprecipitation ChIP',
'meta_title' => 'Chromatin immunoprecipitation ChIP-grade antibodies | Diagenode',
'modified' => '2021-07-01 10:22:38',
'created' => '2017-02-14 11:16:04',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '405',
'name' => 'Datasheet H3K4me3 C15410030',
'description' => '<p>Datasheet description</p>',
'image_id' => null,
'type' => 'Datasheet',
'url' => 'files/products/antibodies/Datasheet_H3K4me3_C15410030.pdf',
'slug' => 'datasheet-h3k4me3-C15410030',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-11-20 17:40:11',
'created' => '2015-07-07 11:47:44',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '11',
'name' => 'Antibodies you can trust',
'description' => '<p style="text-align: justify;"><span>Epigenetic research tools have evolved over time from endpoint PCR to qPCR to the analyses of large sets of genome-wide sequencing data. ChIP sequencing (ChIP-seq) has now become the gold standard method for chromatin studies, given the accuracy and coverage scale of the approach over other methods. Successful ChIP-seq, however, requires a higher level of experimental accuracy and consistency in all steps of ChIP than ever before. Particularly crucial is the quality of ChIP antibodies. </span></p>',
'image_id' => null,
'type' => 'Poster',
'url' => 'files/posters/Antibodies_you_can_trust_Poster.pdf',
'slug' => 'antibodies-you-can-trust-poster',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2015-10-01 20:18:31',
'created' => '2015-07-03 16:05:15',
'ProductsDocument' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1783',
'name' => 'product/antibodies/chipseq-grade-ab-icon.png',
'alt' => 'ChIP-seq Grade',
'modified' => '2020-11-27 07:04:40',
'created' => '2018-03-15 15:54:09',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '4598',
'name' => 'SUMO protease FUG1, histone reader AL3 and the PRC1 Complex areintegral to repeat-expansion induced epigenetic silencing in Arabidopsisthaliana',
'authors' => 'Sureshkumar S. et al.',
'description' => '<p>Epigenetic gene silencing induced by expanded repeats can cause diverse phenotypes ranging from severe growth defects in plants to genetic diseases such as Friedreich’s ataxia in humans1. The molecular mechanisms underlying repeat expansion-induced epigenetic silencing remain largely unknown2,3. Using a plant model, we have previously shown that expanded repeats can induce smallRNAs which in turn can lead to epigenetic silencing through the RNA-dependent DNA methylation pathway4,5. Here, using a genetic suppressor screen, we confirm a key role for the RdDM pathway and identify novel components required for epigenetic silencing caused by expanded repeats. We show that FOURTH ULP LIKE GENE CLASS 1 (FUG1) – a SUMO protease, ALFIN-LIKE 3 – a histone reader and LIKE HETEROCHROMATIN 1 (LHP1) - a component of the PRC1 complex are required for repeat expansion-induced epigenetic silencing. Loss of any of these components suppress repeat expansion-associated phenotypes. SUMO protease FUG1 physically interacts with AL3 and perturbing its potential SUMOylation site disrupts its nuclear localisation. AL3 physically interacts with LHP1 of the PRC1 complex and the FUG1-AL3-LHP1 module is essential to confer repeat expansion-associated epigenetic silencing. Our findings highlight the importance post-translational modifiers and histone readers in epigenetic silencing caused by repeat expansions.</p>',
'date' => '2023-12-01',
'pmid' => 'https://doi.org/10.1101%2F2023.01.13.523841',
'doi' => '10.1101/2023.01.13.523841',
'modified' => '2023-04-06 09:10:33',
'created' => '2023-02-21 09:59:46',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '4692',
'name' => 'Temporal modification of H3K9/14ac and H3K4me3 histone marksmediates mechano-responsive gene expression during the accommodationprocess in poplar',
'authors' => 'Ghosh R. et al.',
'description' => '<p>Plants can attenuate their molecular response to repetitive mechanical stimulation as a function of their mechanical history. For instance, a single bending of stem is sufficient to attenuate the gene expression in poplar plants to the subsequent mechanical stimulation, and the state of desensitization can last for several days. The role of histone modifications in memory gene expression and modulating plant response to abiotic or biotic signals is well known. However, such information is still lacking to explain the attenuated expression pattern of mechano-responsive genes in plants under repetitive stimulation. Using poplar as a model plant in this study, we first measured the global level of H3K9/14ac and H3K4me3 marks in the bent stem. The result shows that a single mild bending of the stem for 6 seconds is sufficient to alter the global level of the H3K9/14ac mark in poplar, highlighting the fact that plants are extremely sensitive to mechanical signals. Next, we analyzed the temporal dynamics of these two active histone marks at attenuated (PtaZFP2, PtaXET6, and PtaACA13) and non-attenuated (PtaHRD) mechano-responsive loci during the desensitization and resensitization phases. Enrichment of H3K9/14ac and H3K4me3 in the regulatory region of attenuated genes correlates well with their transient expression pattern after the first bending. Moreover, the levels of H3K4me3 correlate well with their expression pattern after the second bending at desensitization (3 days after the first bending) as well as resensitization (5 days after the first bending) phases. On the other hand, H3K9/14ac status correlates only with their attenuated expression pattern at the desensitization phase. The expression efficiency of the attenuated genes was restored after the second bending in the histone deacetylase inhibitor-treated plants. While both histone modifications contribute to the expression of attenuated genes, mechanostimulated expression of the non-attenuated PtaHRD gene seems to be H3K4me3 dependent.</p>',
'date' => '2023-02-01',
'pmid' => 'https://doi.org/10.1101%2F2023.02.12.526104',
'doi' => '10.1101/2023.02.12.526104',
'modified' => '2023-04-14 09:20:38',
'created' => '2023-02-28 12:19:11',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '4474',
'name' => 'DNA sequence and chromatin modifiers cooperate to confer epigeneticbistability at imprinting control regions.',
'authors' => 'Butz S. et al.',
'description' => '<p>Genomic imprinting is regulated by parental-specific DNA methylation of imprinting control regions (ICRs). Despite an identical DNA sequence, ICRs can exist in two distinct epigenetic states that are memorized throughout unlimited cell divisions and reset during germline formation. Here, we systematically study the genetic and epigenetic determinants of this epigenetic bistability. By iterative integration of ICRs and related DNA sequences to an ectopic location in the mouse genome, we first identify the DNA sequence features required for maintenance of epigenetic states in embryonic stem cells. The autonomous regulatory properties of ICRs further enabled us to create DNA-methylation-sensitive reporters and to screen for key components involved in regulating their epigenetic memory. Besides DNMT1, UHRF1 and ZFP57, we identify factors that prevent switching from methylated to unmethylated states and show that two of these candidates, ATF7IP and ZMYM2, are important for the stability of DNA and H3K9 methylation at ICRs in embryonic stem cells.</p>',
'date' => '2022-11-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/36333500',
'doi' => '10.1038/s41588-022-01210-z',
'modified' => '2022-11-18 12:20:16',
'created' => '2022-11-15 09:26:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '4345',
'name' => 'Altered Chromatin States Drive Cryptic Transcription in AgingMammalian Stem Cells.',
'authors' => 'McCauley Brenna S et al.',
'description' => '<p>A repressive chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation suppresses cryptic transcription in embryonic stem cells. Cryptic transcription is elevated with age in yeast and nematodes, and reducing it extends yeast lifespan, though whether this occurs in mammals is unknown. We show that cryptic transcription is elevated in aged mammalian stem cells, including murine hematopoietic stem cells (mHSCs) and neural stem cells (NSCs) and human mesenchymal stem cells (hMSCs). Precise mapping allowed quantification of age-associated cryptic transcription in hMSCs aged . Regions with significant age-associated cryptic transcription have a unique chromatin signature: decreased H3K36me3 and increased H3K4me1, H3K4me3, and H3K27ac with age. Genomic regions undergoing such changes resemble known promoter sequences and are bound by TBP even in young cells. Hence, the more permissive chromatin state at intragenic cryptic promoters likely underlies increased cryptic transcription in aged mammalian stem cells.</p>',
'date' => '2021-08-01',
'pmid' => 'https://doi.org/10.1038%2Fs43587-021-00091-x',
'doi' => '10.1038/s43587-021-00091-x',
'modified' => '2022-06-22 12:30:19',
'created' => '2022-05-19 10:41:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '4384',
'name' => 'Age-associated cryptic transcription in mammalian stem cells is linked topermissive chromatin at cryptic promoters',
'authors' => 'McCauley B. S. et al.',
'description' => '<p>Suppressing spurious cryptic transcription by a repressive intragenic chromatin state featuring trimethylated lysine 36 on histone H3 (H3K36me3) and DNA methylation is critical for maintaining self-renewal capacity in mouse embryonic stem cells. In yeast and nematodes, such cryptic transcription is elevated with age, and reducing the levels of age-associated cryptic transcription extends yeast lifespan. Whether cryptic transcription is also increased during mammalian aging is unknown. We show for the first time an age-associated elevation in cryptic transcription in several stem cell populations, including murine hematopoietic stem cells (mHSCs) and neural stem cells (NSCs) and human mesenchymal stem cells (hMSCs). Using DECAP-seq, we mapped and quantified age-associated cryptic transcription in hMSCs aged in vitro. Regions with significant age-associated cryptic transcription have a unique chromatin signature: decreased H3K36me3 and increased H3K4me1, H3K4me3, and H3K27ac with age. Furthermore, genomic regions undergoing such age-dependent chromatin changes resemble known promoter sequences and are bound by the promoter-associated protein TBP even in young cells. Hence, the more permissive chromatin state at intragenic cryptic promoters likely underlies the increase of cryptic transcription in aged mammalian stem cells.</p>',
'date' => '2020-10-01',
'pmid' => 'https://europepmc.org/article/ppr/ppr221829',
'doi' => '10.21203/rs.3.rs-82156/v1',
'modified' => '2022-08-04 16:24:46',
'created' => '2022-08-04 14:55:36',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '3990',
'name' => 'Identification of Novel Molecular Markers of Human Th17 Cells.',
'authors' => 'Sałkowska A, Karaś K, Karwaciak I, Walczak-Drzewiecka A, Krawczyk M, Sobalska-Kwapis M, Dastych J, Ratajewski M',
'description' => '<p>Th17 cells are important players in host defense against pathogens such as , , and . Th17 cell-mediated inflammation, under certain conditions in which balance in the immune system is disrupted, is the underlying pathogenic mechanism of certain autoimmune disorders, e.g., rheumatoid arthritis, Graves' disease, multiple sclerosis, and psoriasis. In the present study, using transcriptomic profiling, we selected genes and analyzed the expression of these genes to find potential novel markers of Th17 lymphocytes. We found that (apolipoprotein D); (complement component 1, Q subcomponent-like protein 1); and (cathepsin L) are expressed at significantly higher mRNA and protein levels in Th17 cells than in the Th1, Th2, and Treg subtypes. Interestingly, these genes and the proteins they encode are well associated with the function of Th17 cells, as these cells produce inflammation, which is linked with atherosclerosis and angiogenesis. Furthermore, we found that high expression of these genes in Th17 cells is associated with the acetylation of H2BK12 within their promoters. Thus, our results provide new information regarding this cell type. Based on these results, we also hope to better identify pathological conditions of clinical significance caused by Th17 cells.</p>',
'date' => '2020-07-03',
'pmid' => 'http://www.pubmed.gov/32635226',
'doi' => '10.3390/cells9071611',
'modified' => '2020-09-01 15:05:28',
'created' => '2020-08-21 16:41:39',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '3695',
'name' => 'The Chromatin Factor HNI9 and ELONGATED HYPOCOTYL5 Maintain ROS Homeostasis under High Nitrogen Provision.',
'authors' => 'Bellegarde F, Maghiaoui A, Boucherez J, Krouk G, Lejay L, Bach L, Gojon A, Martin A',
'description' => '<p>Reactive oxygen species (ROS) can accumulate in cells at excessive levels, leading to unbalanced redox states and to potential oxidative stress, which can have damaging effects on the molecular components of plant cells. Several environmental conditions have been described as causing an elevation of ROS production in plants. Consequently, activation of detoxification responses is necessary to maintain ROS homeostasis at physiological levels. Misregulation of detoxification systems during oxidative stress can ultimately cause growth retardation and developmental defects. Here, we demonstrate that Arabidopsis () plants grown in a high nitrogen (N) environment express a set of genes involved in detoxification of ROS that maintain ROS at physiological levels. We show that the chromatin factor HIGH NITROGEN INSENSITIVE9 (HNI9) is an important mediator of this response and is required for the expression of detoxification genes. Mutation in HNI9 leads to elevated ROS levels and ROS-dependent phenotypic defects under high but not low N provision. In addition, we identify ELONGATED HYPOCOTYL5 as a major transcription factor required for activation of the detoxification program under high N. Our results demonstrate the requirement of a balance between N metabolism and ROS production, and our work establishes major regulators required to control ROS homeostasis under conditions of excess N.</p>',
'date' => '2019-05-01',
'pmid' => 'http://www.pubmed.gov/30824566',
'doi' => '10.1104/pp.18.01473',
'modified' => '2019-06-28 13:46:23',
'created' => '2019-06-21 14:55:31',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '3579',
'name' => 'Polycomb Repressive Complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1.',
'authors' => 'Bellegarde F, Herbert L, Séré D, Caillieux E, Boucherez J, Fizames C, Roudier F, Gojon A, Martin A',
'description' => '<p>PRC2 is a major regulator of gene expression in eukaryotes. It catalyzes the repressive chromatin mark H3K27me3, which leads to very low expression of target genes. NRT2.1, which encodes a key root nitrate transporter in Arabidopsis, is targeted by H3K27me3, but the function of PRC2 on NRT2.1 remains unclear. Here, we demonstrate that PRC2 directly targets and down-regulates NRT2.1, but in a context of very high transcription, in nutritional conditions where this gene is one of the most highly expressed genes in the transcriptome. Indeed, the mutation of CLF, which encodes a PRC2 subunit, leads to a loss of H3K27me3 at NRT2.1 and results, exclusively under permissive conditions for NRT2.1, in a further increase in NRT2.1 expression, and specifically in tissues where NRT2.1 is normally expressed. Therefore, our data indicates that PRC2 tempers the hyperactivity of NRT2.1 in a context of very strong transcription. This reveals an original function of PRC2 in the control of the expression of a highly expressed gene in Arabidopsis.</p>',
'date' => '2018-05-21',
'pmid' => 'http://www.pubmed.gov/29784958',
'doi' => '10.1038/s41598-018-26349-w',
'modified' => '2019-04-17 15:55:09',
'created' => '2019-04-16 12:25:30',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '3377',
'name' => 'Interplay of cell–cell contacts and RhoA/MRTF‐A signaling regulates cardiomyocyte identity',
'authors' => 'Dorn et al',
'description' => '<p><span>Cell–cell and cell–matrix interactions guide organ development and homeostasis by controlling lineage specification and maintenance, but the underlying molecular principles are largely unknown. Here, we show that in human developing cardiomyocytes cell–cell contacts at the intercalated disk connect to remodeling of the actin cytoskeleton by regulating the RhoA‐</span><span class="styled-content fixed-case">ROCK</span><span><span> </span>signaling to maintain an active<span> </span></span><span class="styled-content fixed-case">MRTF</span><span>/</span><span class="styled-content fixed-case">SRF</span><span><span> </span>transcriptional program essential for cardiomyocyte identity. Genetic perturbation of this mechanosensory pathway activates an ectopic fat gene program during cardiomyocyte differentiation, which ultimately primes the cells to switch to the brown/beige adipocyte lineage in response to adipogenesis‐inducing signals. We also demonstrate by<span> </span></span><em>in vivo</em><span><span> </span>fate mapping and clonal analysis of cardiac progenitors that cardiac fat and a subset of cardiac muscle arise from a common precursor expressing Isl1 and Wt1 during heart development, suggesting related mechanisms of determination between the two lineages.</span></p>',
'date' => '2018-05-15',
'pmid' => 'http://emboj.embopress.org/content/early/2018/05/15/embj.201798133',
'doi' => '10.15252/embj.201798133',
'modified' => '2018-05-22 22:50:53',
'created' => '2018-05-22 22:50:53',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '3473',
'name' => 'Neonatal exposure to hyperoxia leads to persistent disturbances in pulmonary histone signatures associated with NOS3 and STAT3 in a mouse model.',
'authors' => 'Chao CM, van den Bruck R, Lork S, Merkle J, Krampen L, Weil PP, Aydin M, Bellusci S, Jenke AC, Postberg J',
'description' => '<p>Background: Early pulmonary oxygen exposure is one of the most important factors implicated in the development of bronchopulmonary dysplasia (BPD). Methods: Here, we analyzed short- and long-term effects of neonatal hyperoxia on NOS3 and STAT3 expression and corresponding epigenetic signatures using a hyperoxia-based mouse model of BPD. Results: Early hyperoxia exposure led to a significant increase in NOS3 (median fold change × 2.37, IQR 1.54-3.68) and STAT3 (median fold change × 2.83, IQR 2.21-3.88) mRNA levels in pulmonary endothelial cells with corresponding changes in histone modification patterns such as H2aZac and H3K9ac hyperacetylation at the respective gene loci. No complete restoration in histone signatures at these loci was observed, and responsivity to later hyperoxia was altered in mouse lungs. In vitro, histone signatures in human aortic endothelial cells (HAEC) remained altered for several weeks after an initial long-term exposure to trichostatin A. This was associated with a substantial increase in baseline eNOS (median 27.2, IQR 22.3-35.6) and STAT3α (median 5.8, IQR 4.8-7.3) mRNA levels with a subsequent significant reduction in eNOS expression upon exposure to hypoxia. Conclusions: Early hyperoxia induced permanent changes in histones signatures at the NOS3 and STAT3 gene locus might partly explain the altered vascular response patterns in children with BPD.</p>',
'date' => '2018-01-01',
'pmid' => 'http://www.pubmed.gov/29581793',
'doi' => '10.1186/s13148-018-0469-0',
'modified' => '2019-02-15 21:45:12',
'created' => '2019-02-14 15:01:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '3299',
'name' => 'Rapid Communication: The correlation between histone modifications and expression of key genes involved in accumulation of adipose tissue in the pig.',
'authors' => 'Kociucka B. et al.',
'description' => '<p>Histone modification is a well-known epigenetic mechanism involved in regulation of gene expression; however, it has been poorly studied in adipose tissues of the pig. Understanding the molecular background of adipose tissue development and function is essential for improving production efficiency and meat quality. The objective of this study was to identify the association between histone modification and the transcript level of genes important for lipid droplet formation and metabolism. Histone modifications at the promoter regions of 6 genes (, , , , , and ) were analyzed using a chromatin immunoprecipitation assay. Two modifications involved in activation of gene expression (acetylation of H3 histone at lysine 9 and methylation of H3 histone at lysine 4) as well as methylation of H3 histone at lysine 27, which is known to be related to gene repression, were examined. The level of histone modification was compared with transcript abundance determined using real-time PCR in tissue samples (subcutaneous fat, visceral fat, and longissimus dorsi muscle) derived from 3 pig breeds significantly differing in fatness traits (Polish Large White, Duroc, and Pietrain). Transcript levels were found to be correlated with histone modifications characteristic to active loci in 4 of 6 genes. A positive correlation between histone H3 lysine 9 acetylation modification and the transcript level of ( = 0.53, < 4.8 × 10), ( = 0.34, < 0.02), and ( = 0.43, < 1.0 × 10) genes was observed. The histone H3 lysine 4 trimethylation modification correlated with transcripts of ( = 0.64, < 4.6 × 10) and ( = 0.37, < 0.01) genes. No correlation was found between transcript level of all studied genes and histone H3 lysine 27 trimethylation level. This is the first study on histone modifications in porcine adipose tissues. We confirmed the relationship between histone modifications and expression of key genes for adipose tissue accumulation in the pig. Epigenetic modulation of the transcriptional profile of these genes (e.g., through nutritional factors) may improve porcine fatness traits in future.</p>',
'date' => '2017-10-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/29108067',
'doi' => '',
'modified' => '2017-12-05 10:39:56',
'created' => '2017-12-05 09:31:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '3083',
'name' => 'Lhx2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11',
'authors' => 'Muralidharan B. et al.',
'description' => '<p>n the developing cerebral cortex, sequential transcriptional programs take neuroepithelial cells from proliferating progenitors to differentiated neurons with unique molecular identities. The regulatory changes that occur in the chromatin of the progenitors are not well understood. During deep layer neurogenesis, we show that transcription factor Lhx2 binds to distal regulatory elements of Fezf2 and Sox11, critical determinants of neuron subtype identity in the mouse neocortex. We demonstrate that Lhx2 binds to the NuRD histone remodeling complex subunits LSD1, HDAC2, and RBBP4, which are proximal regulators of the epigenetic state of chromatin. When Lhx2 is absent, active histone marks at the Fezf2 and Sox11 loci are increased. Loss of Lhx2 produces an increase, and overexpression of Lhx2 causes a decrease, in layer 5 Fezf2 and Ctip2 expressing neurons. Our results provide mechanistic insight into how Lhx2 acts as a necessary and sufficient regulator of genes that control cortical neuronal subtype identity.</p>',
'date' => '2016-12-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/27909100',
'doi' => '',
'modified' => '2016-12-20 10:28:02',
'created' => '2016-12-20 10:28:02',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '2910',
'name' => 'Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape',
'authors' => 'Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G',
'description' => '<div class="">
<h4>BACKGROUND:</h4>
<p><abstracttext label="BACKGROUND" nlmcategory="BACKGROUND">Previously, a role was demonstrated for transcription in the acquisition of DNA methylation at imprinted control regions in oocytes. Definition of the oocyte DNA methylome by whole genome approaches revealed that the majority of methylated CpG islands are intragenic and gene bodies are hypermethylated. Yet, the mechanisms by which transcription regulates DNA methylation in oocytes remain unclear. Here, we systematically test the link between transcription and the methylome.</abstracttext></p>
<h4>RESULTS:</h4>
<p><abstracttext label="RESULTS" nlmcategory="RESULTS">We perform deep RNA-Seq and de novo transcriptome assembly at different stages of mouse oogenesis. This reveals thousands of novel non-annotated genes, as well as alternative promoters, for approximately 10 % of reference genes expressed in oocytes. In addition, a large fraction of novel promoters coincide with MaLR and ERVK transposable elements. Integration with our transcriptome assembly reveals that transcription correlates accurately with DNA methylation and accounts for approximately 85-90 % of the methylome. We generate a mouse model in which transcription across the Zac1/Plagl1 locus is abrogated in oocytes, resulting in failure of DNA methylation establishment at all CpGs of this locus. ChIP analysis in oocytes reveals H3K4me2 enrichment at the Zac1 imprinted control region when transcription is ablated, establishing a connection between transcription and chromatin remodeling at CpG islands by histone demethylases.</abstracttext></p>
<h4>CONCLUSIONS:</h4>
<p><abstracttext label="CONCLUSIONS" nlmcategory="CONCLUSIONS">By precisely defining the mouse oocyte transcriptome, this work not only highlights transcription as a cornerstone of DNA methylation establishment in female germ cells, but also provides an important resource for developmental biology research.</abstracttext></p>
</div>',
'date' => '2015-09-25',
'pmid' => 'http://www.ncbi.nlm.nih.gov/pubmed/26408185',
'doi' => '10.1186/s13059-015-0769-z',
'modified' => '2016-05-10 22:01:49',
'created' => '2016-05-10 22:01:49',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '2613',
'name' => 'CpG signalling, H2A.Z/H3 acetylation and microRNA-mediated deferred self-attenuation orchestrate foetal NOS3 expression.',
'authors' => 'Postberg J, Kanders M, Forcob S, Willems R, Orth V, Hensel KO, Weil PP, Wirth S, Jenke AC',
'description' => 'BACKGROUND: An adverse intrauterine environment leads to permanent physiological changes including vascular tone regulation, potentially influencing the risk for adult vascular diseases. We therefore aimed to monitor responsive NOS3 expression in human umbilical artery endothelial cells (HUAEC) and to study the underlying epigenetic signatures involved in its regulation. RESULTS: NOS3 and STAT3 mRNA levels were elevated in HUAEC of patients who suffered from placental insufficiency. 5-hydroxymethylcytosine, H3K9ac and Histone 2A (H2A).Zac at the NOS3 transcription start site directly correlated with NOS3 mRNA levels. Concomitantly, we observed entangled histone acetylation patterns and NOS3 response upon hypoxic conditions in vitro. Knock-down of either NOS3 or STAT3 by RNAi provided evidence for a functional NOS3/STAT3 relationship. Moreover, we recognized massive turnover of Stat3 at a discrete binding site in the NOS3 promoter. Interestingly, induced hyperacetylation resulted in short-termed increase of NOS3 mRNA followed by deferred decrease indicating that NOS3 expression could become self-attenuated by co-expressed intronic 27 nt-ncRNA. Reporter assay results and phylogenetic analyses enabled us to propose a novel model for STAT3-3'-UTR targeting by this 27-nt-ncRNA. CONCLUSIONS: An adverse intrauterine environment leads to adaptive changes of NOS3 expression. Apparently, a rapid NOS3 self-limiting response upon ectopic triggers co-exists with longer termed expression changes in response to placental insufficiency involving differential epigenetic signatures. Their persistence might contribute to impaired vascular endothelial response and consequently increase the risk of cardiovascular disease later in life.',
'date' => '2015-01-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25699114',
'doi' => '',
'modified' => '2015-07-24 15:39:05',
'created' => '2015-07-24 15:39:05',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '2506',
'name' => 'Paclitaxel resistance increases oncolytic adenovirus efficacy via upregulated CAR expression and dysfunctional cell cycle control.',
'authors' => 'Ingemarsdotter CK, Tookman LA, Browne A, Pirlo K, Cutts R, Chelela C, Khurrum KF, Leung EY, Dowson S, Webber L, Khan I, Ennis D, Syed N, Crook TR, Brenton JD, Lockley M, McNeish IA',
'description' => 'Resistance to paclitaxel chemotherapy frequently develops in ovarian cancer. Oncolytic adenoviruses are a novel therapy for human malignancies that are being evaluated in early phase trials. However, there are no reliable predictive biomarkers for oncolytic adenovirus activity in ovarian cancer. We investigated the link between paclitaxel resistance and oncolytic adenovirus activity using established ovarian cancer cell line models, xenografts with de novo paclitaxel resistance and tumour samples from two separate trials. The activity of multiple Ad5 vectors, including dl922-947 (E1A CR2-deleted), dl1520 (E1B-55K deleted) and Ad5 WT, was significantly increased in paclitaxel resistant ovarian cancer in vitro and in vivo. This was associated with greater infectivity resulting from increased expression of the primary receptor for Ad5, CAR (coxsackie adenovirus receptor). This, in turn, resulted from increased CAR transcription secondary to histone modification in resistant cells. There was increased CAR expression in intraperitoneal tumours with de novo paclitaxel resistance and in tumours from patients with clinical resistance to paclitaxel. Increased CAR expression did not cause paclitaxel resistance, but did increase inflammatory cytokine expression. Finally, we identified dysregulated cell cycle control as a second mechanism of increased adenovirus efficacy in paclitaxel-resistant ovarian cancer. Ad11 and Ad35, both group B adenoviruses that utilise non-CAR receptors to infect cells, are also significantly more effective in paclitaxel-resistant ovarian cell models. Inhibition of CDK4/6 using PD-0332991 was able both to reverse paclitaxel resistance and reduce adenovirus efficacy. Thus, paclitaxel resistance increases oncolytic adenovirus efficacy via at least two separate mechanisms - if validated further, this information could have future clinical utility to aid patient selection for clinical trials.',
'date' => '2014-12-29',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/25560085',
'doi' => '',
'modified' => '2015-07-24 15:39:04',
'created' => '2015-07-24 15:39:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '998',
'name' => 'p53-Independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6.',
'authors' => 'Phalke S, Mzoughi S, Bezzi M, Jennifer N, Mok WC, Low DH, Thike AA, Kuznetsov VA, Tan PH, Voorhoeve PM, Guccione E',
'description' => 'p21 is a potent cyclin-dependent kinase inhibitor that plays a role in promoting G1 cell cycle arrest and cellular senescence. Consistent with this role, p21 is a downstream target of several tumour suppressors and oncogenes, and it is downregulated in the majority of tumours, including breast cancer. Here, we report that protein arginine methyltransferase 6 (PRMT6), a type I PRMT known to act as a transcriptional cofactor, directly represses the p21 promoter. PRMT6 knock-down (KD) results in a p21 derepression in breast cancer cells, which is p53-independent, and leads to cell cycle arrest, cellular senescence and reduced growth in soft agar assays and in severe combined immunodeficiency (SCID) mice for all the cancer lines examined. We finally show that bypassing the p21-mediated arrest rescues PRMT6 KD cells from senescence, and it restores their ability to grow on soft agar. We conclude that PRMT6 acts as an oncogene in breast cancer cells, promoting growth and preventing senescence, making it an attractive target for cancer therapy.',
'date' => '2012-09-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22987071',
'doi' => '',
'modified' => '2015-07-24 15:38:59',
'created' => '2015-07-24 15:38:59',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '359',
'name' => 'Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.',
'authors' => 'Henckel A, Chebli K, Kota SK, Arnaud P, Feil R',
'description' => 'Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at 'maternal' ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.',
'date' => '2012-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22117218',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '432',
'name' => 'H3K4me3 antibody SDS US en',
'language' => 'en',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-US-en-GHS_2_0.pdf',
'countries' => 'US',
'modified' => '2020-06-30 18:23:32',
'created' => '2020-06-30 18:23:32',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '430',
'name' => 'H3K4me3 antibody SDS GB en',
'language' => 'en',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-GB-en-GHS_2_0.pdf',
'countries' => 'GB',
'modified' => '2020-06-30 18:22:36',
'created' => '2020-06-30 18:22:36',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '425',
'name' => 'H3K4me3 antibody SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-fr-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:17:31',
'created' => '2020-06-30 18:17:31',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '429',
'name' => 'H3K4me3 antibody SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-FR-fr-GHS_2_0.pdf',
'countries' => 'FR',
'modified' => '2020-06-30 18:22:02',
'created' => '2020-06-30 18:22:02',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '428',
'name' => 'H3K4me3 antibody SDS ES es',
'language' => 'es',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-ES-es-GHS_2_0.pdf',
'countries' => 'ES',
'modified' => '2020-06-30 18:21:17',
'created' => '2020-06-30 18:21:17',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '427',
'name' => 'H3K4me3 antibody SDS DE de',
'language' => 'de',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-DE-de-GHS_2_0.pdf',
'countries' => 'DE',
'modified' => '2020-06-30 18:19:09',
'created' => '2020-06-30 18:19:09',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '431',
'name' => 'H3K4me3 antibody SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-JP-ja-GHS_2_0.pdf',
'countries' => 'JP',
'modified' => '2020-06-30 18:23:02',
'created' => '2020-06-30 18:23:02',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '426',
'name' => 'H3K4me3 antibody SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:18:22',
'created' => '2020-06-30 18:18:22',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://www.diagenode.com/en/p/h3k4me3-polyclonal-antibody-classic-50-ug-25-ul'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
)
$pro = array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = '<li>
<div class="row">
<div class="small-12 columns">
<a href="/en/p/bioruptor-pico-sonication-device"><img src="/img/product/shearing_technologies/bioruptor_pico.jpg" alt="Bioruptor pico next gen sequencing " class="th"/></a> </div>
<div class="small-12 columns">
<div class="small-6 columns" style="padding-left:0px;padding-right:0px;margin-top:-6px;margin-left:-1px">
<span class="success label" style="">B01060010</span>
</div>
<div class="small-6 columns text-right" style="padding-left:0px;padding-right:0px;margin-top:-6px">
<!--a href="#" style="color:#B21329"><i class="fa fa-info-circle"></i></a-->
</div>
</div>
<div class="small-12 columns" >
<h6 style="height:60px">Bioruptor® Pico sonication device</h6>
</div>
</div>
</li>
'
$related = array(
'id' => '1787',
'antibody_id' => null,
'name' => 'Bioruptor<sup>®</sup> Pico sonication device',
'description' => '<p><a href="https://go.diagenode.com/bioruptor-upgrade"><img src="https://www.diagenode.com/img/banners/banner-br-trade.png" /></a></p>
<p>The Bioruptor® Pico (2013-2019) represented a breakthrough for shearing micro-volumes of 5 μl to larger volumes of up to 2 ml. <span>The new generation keeps the features you like the most and bring even more innovation. Check it now:</span></p>
<center><span></span></center><center><a href="https://www.diagenode.com/p/bioruptorpico2"> <img alt="New Bioruptor Pico" src="https://www.diagenode.com/img/product/shearing_technologies/new-pico-product-banner.jpg" /></a></center>
<p></p>
<p><span>Watch our short video about the Bioruptor Pico and how it can help you accomplish perfect shearing for any application including chromatin shearing, DNA shearing for NGS, unmatched DNA extraction from FFPE samples, RNA shearing, protein extraction, and much more.</span></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<center><iframe width="560" height="315" src="https://www.youtube.com/embed/ckLc4owudIM" frameborder="0" allowfullscreen="allowfullscreen"></iframe></center><center>
<p></p>
</center><center><a href="https://www.diagenode.com/en/pages/osha"><img src="https://www.diagenode.com/img/banners/banner-osha-580.jpg" width="635" height="243" /></a></center>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label1' => 'User manual ',
'info1' => '<p><a href="https://www.diagenode.com/files/products/shearing_technology/bioruptor/Bioruptor_pico_cooler_manual.pdf">Download</a></p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label2' => 'Recommended settings for DNA shearing with Bioruptor® Pico',
'info2' => '<p>Follow our guidelines and find the good parameters for your expected DNA size: <a href="https://pybrevet.typeform.com/to/o8cQfM">DNA shearing with the Bioruptor® Pico</a></p>
<p></p>
<p>
<script>// <![CDATA[
(function(){var qs,js,q,s,d=document,gi=d.getElementById,ce=d.createElement,gt=d.getElementsByTagName,id='typef_orm',b='https://s3-eu-west-1.amazonaws.com/share.typeform.com/';if(!gi.call(d,id)){js=ce.call(d,'script');js.id=id;js.src=b+'share.js';q=gt.call(d,'script')[0];q.parentNode.insertBefore(js,q)}id=id+'_';if(!gi.call(d,id)){qs=ce.call(d,'link');qs.rel='stylesheet';qs.id=id;qs.href=b+'share-button.css';s=gt.call(d,'head')[0];s.appendChild(qs,s)}})()
// ]]></script>
</p>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'label3' => 'Available chromatin shearing kits',
'info3' => '<p>It is important to establish optimal conditions to shear crosslinked chromatin to get the correct fragment sizes needed for ChIP. Usually this process requires both optimizing sonication conditions as well as optimizing SDS concentration, which is laborious. With the Chromatin Shearing Optimization Kits, optimization is fast and easy - we provide optimization reagents with varying concentrations of SDS. Moreover, our Chromatin Shearing Optimization Kits can be used for the optimization of chromatin preparation with our kits for ChIP.</p>
<table style="width: 925px;">
<tbody>
<tr valign="middle">
<td style="width: 213px;"></td>
<td style="text-align: center; width: 208px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-100-million-cells">Chromatin Shearing Kit Low SDS (for Histones)</a></strong></td>
<td style="text-align: center; width: 180px;"><strong><a href="../p/chromatin-shearing-optimization-kit-low-sds-for-tfs-25-rxns">Chromatin Shearing Kit Low SDS (for TF)</a></strong></td>
<td style="text-align: center; width: 154px;"><strong><a href="../p/chromatin-shearing-optimization-kit-high-sds-100-million-cells">Chromatin Shearing Kit High SDS</a></strong></td>
<td style="text-align: center; width: 155px;"><strong><a href="../p/chromatin-shearing-plant-chip-seq-kit">Chromatin Shearing Kit (for Plant)</a></strong></td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>SDS concentration</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">< 0.1%</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">0.2%</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">1%</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">0.5%</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Nuclei isolation</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">Yes</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">No</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">Yes</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Allows for shearing of... cells/tissue</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;">100 million cells</p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;">up to 25 g of tissue</p>
</td>
</tr>
<tr style="background-color: #fff;" valign="middle">
<td style="width: 213px;">
<p style="text-align: left;"><strong>Corresponding to shearing buffers from</strong></p>
</td>
<td style="text-align: center; width: 208px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-x24-24-rxns">iDeal ChIP-seq kit for Histones</a></p>
<p style="text-align: center;"><a href="https://www.diagenode.com/en/p/manual-chipmentation-kit-for-histones-24-rxns">ChIPmentation Kit for Histones</a></p>
</td>
<td style="text-align: center; width: 180px;">
<p style="text-align: center;"><a href="../p/ideal-chip-seq-kit-for-transcription-factors-x24-24-rxns">iDeal ChIP-seq Kit for Transcription Factors</a></p>
<p style="text-align: center;"><a href="../p/ideal-chip-qpcr-kit">iDeal ChIP qPCR kit</a></p>
</td>
<td style="text-align: center; width: 154px;">
<p style="text-align: center;"><a href="../p/true-microchip-kit-x16-16-rxns">True MicroChIP kit</a></p>
</td>
<td style="text-align: center; width: 155px;">
<p style="text-align: center;"><a href="../p/universal-plant-chip-seq-kit-x24-24-rxns">Universal Plant <br />ChIP-seq kit</a></p>
</td>
</tr>
</tbody>
</table>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<script src="chrome-extension://hhojmcideegachlhfgfdhailpfhgknjm/web_accessible_resources/index.js"></script>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>
<div id="ConnectiveDocSignExtentionInstalled" data-extension-version="1.0.4"></div>',
'format' => '1 unit',
'catalog_number' => 'B01060010',
'old_catalog_number' => '',
'sf_code' => 'B01060010-',
'type' => 'ACC',
'search_order' => '00-Machine',
'price_EUR' => '24000',
'price_USD' => '27500',
'price_GBP' => '21000',
'price_JPY' => '3759600',
'price_CNY' => 'Discontinued',
'price_AUD' => '68750',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => true,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '0000-00-00',
'slug' => 'bioruptor-pico-sonication-device',
'meta_title' => 'Bioruptor® Pico sonication device for RNA,Chromatin and DNA shearing for Next-Generation-Sequencing | Diagenode',
'meta_keywords' => 'Bioruptor, sonication, Next-Generation-Sequencing,DNA shearing,Protein extraction',
'meta_description' => 'An all-in-one shearing system Ideal for DNA shearing for Next-Generation-Sequencing,Chromatin shearing,RNA shearing,Protein extraction from tissues and cells and FFPE DNA extraction',
'modified' => '2023-12-20 14:21:02',
'created' => '2015-06-29 14:08:20',
'ProductsRelated' => array(
'id' => '1101',
'product_id' => '2195',
'related_id' => '1787'
),
'Image' => array(
(int) 0 => array(
'id' => '133',
'name' => 'product/shearing_technologies/bioruptor_pico.jpg',
'alt' => 'Bioruptor pico next gen sequencing ',
'modified' => '2020-11-27 10:00:30',
'created' => '2015-06-10 17:28:55',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '1772',
'name' => 'product/shearing_technologies/B010600010.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 15:41:46',
'created' => '2018-02-14 15:41:46',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '1773',
'name' => 'product/shearing_technologies/B010600010-2.jpg',
'alt' => 'B010600010',
'modified' => '2018-02-14 16:18:03',
'created' => '2018-02-14 15:42:00',
'ProductsImage' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '1774',
'name' => 'product/shearing_technologies/B01060010-bioruptor-pico-cooler.jpg',
'alt' => 'Pico + cooler',
'modified' => '2018-03-06 11:01:33',
'created' => '2018-03-06 11:01:33',
'ProductsImage' => array(
[maximum depth reached]
)
)
)
)
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '3143',
'antibody_id' => '617',
'name' => 'H3K4me3 Antibody (sample size)',
'description' => '',
'label1' => '',
'info1' => '',
'label2' => '',
'info2' => '',
'label3' => '',
'info3' => '',
'format' => '10 µg',
'catalog_number' => 'C15410030-10',
'old_catalog_number' => '',
'sf_code' => 'C15410030-D001-000582',
'type' => 'FRE',
'search_order' => '03-Antibody',
'price_EUR' => '105',
'price_USD' => '115',
'price_GBP' => '100',
'price_JPY' => '16450',
'price_CNY' => '',
'price_AUD' => '288',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => 'May 3, 2018',
'slug' => 'h3k4me3-polyclonal-antibody-classic-10',
'meta_title' => '',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-12-23 11:31:53',
'created' => '2020-11-25 16:06:28',
'ProductsGroup' => array(
'id' => '362',
'product_id' => '3143',
'group_id' => '322'
)
)
$img = 'banners/banner-cut_tag-chipmentation-500.jpg'
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$application = array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'ProductsApplication' => array(
'id' => '4314',
'product_id' => '2195',
'application_id' => '42'
)
)
$slugs = array(
(int) 0 => 'chip-seq-antibodies'
)
$applications = array(
'id' => '42',
'position' => '10',
'parent_id' => '40',
'name' => 'ChIP-seq (ab)',
'description' => '',
'in_footer' => false,
'in_menu' => false,
'online' => true,
'tabular' => true,
'slug' => 'chip-seq-antibodies',
'meta_keywords' => 'Chromatin Immunoprecipitation Sequencing,ChIP-Seq,ChIP-seq grade antibodies,DNA purification,qPCR,Shearing of chromatin',
'meta_description' => 'Diagenode offers a wide range of antibodies and technical support for ChIP Sequencing applications',
'meta_title' => 'ChIP Sequencing Antibodies (ChIP-Seq) | Diagenode',
'modified' => '2016-01-20 11:06:19',
'created' => '2015-10-20 11:44:45',
'locale' => 'eng'
)
$description = ''
$name = 'ChIP-seq (ab)'
$document = array(
'id' => '38',
'name' => 'Epigenetic Antibodies Brochure',
'description' => '<p>More than in any other immuoprecipitation assays, quality antibodies are critical tools in many epigenetics experiments. Since 10 years, Diagenode has developed the most stringent quality production available on the market for antibodies exclusively focused on epigenetic uses. All our antibodies have been qualified to work in epigenetic applications.</p>',
'image_id' => null,
'type' => 'Brochure',
'url' => 'files/brochures/Epigenetic_Antibodies_Brochure.pdf',
'slug' => 'epigenetic-antibodies-brochure',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2016-06-15 11:24:06',
'created' => '2015-07-03 16:05:27',
'ProductsDocument' => array(
'id' => '1616',
'product_id' => '2195',
'document_id' => '38'
)
)
$sds = array(
'id' => '426',
'name' => 'H3K4me3 antibody SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/H3K4me3/SDS-C15410030-H3K4me3_Antibody-BE-nl-GHS_2_0.pdf',
'countries' => 'BE',
'modified' => '2020-06-30 18:18:22',
'created' => '2020-06-30 18:18:22',
'ProductsSafetySheet' => array(
'id' => '833',
'product_id' => '2195',
'safety_sheet_id' => '426'
)
)
$publication = array(
'id' => '359',
'name' => 'Transcription and histone methylation changes correlate with imprint acquisition in male germ cells.',
'authors' => 'Henckel A, Chebli K, Kota SK, Arnaud P, Feil R',
'description' => 'Genomic imprinting in mammals is controlled by DNA methylation imprints that are acquired in the gametes, at essential sequence elements called 'imprinting control regions' (ICRs). What signals paternal imprint acquisition in male germ cells remains unknown. To address this question, we explored histone methylation at ICRs in mouse primordial germ cells (PGCs). By 13.5 days post coitum (d.p.c.), H3 lysine-9 and H4 lysine-20 trimethylation are depleted from ICRs in male (and female) PGCs, indicating that these modifications do not signal subsequent imprint acquisition, which initiates at ∼15.5 d.p.c. Furthermore, during male PGC development, H3 lysine-4 trimethylation becomes biallelically enriched at 'maternal' ICRs, which are protected against DNA methylation, and whose promoters are active in the male germ cells. Remarkably, high transcriptional read-through is detected at the paternal ICRs H19-DMR and Ig-DMR at the time of imprint establishment, from one of the strands predominantly. Combined, our data evoke a model in which differential histone modification states linked to transcriptional events may signal the specificity of imprint acquisition during spermatogenesis.',
'date' => '2012-02-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/22117218',
'doi' => '',
'modified' => '2015-07-24 15:38:57',
'created' => '2015-07-24 15:38:57',
'ProductsPublication' => array(
'id' => '870',
'product_id' => '2195',
'publication_id' => '359'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/22117218" target="_blank"><i class="fa fa-external-link"></i></a>'include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1052
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
×